Gene
KWMTBOMO10207
Pre Gene Modal
BGIBMGA005604
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.981
Sequence
CDS
ATGGAGAGTAACAGAAAAGTGCAGTATCTGGCCGCATTTAGTGTGTCTTTAGCTTTCTTAGCGGTGGGAGCGTGCTCGGCATGGCCCACACCCGTATTACTGAAATTTCATAATAATGAAACTTCCGTGGTGATTTCCAATACGGAGATATCATGGATGCTAGGACTGAACCCAATTGGATTCCTTATCGGCAGTTTCACGGCACGCTTCGTTATAGACAAGTTTGGGAGAAGATATACATTATTAGCTAGTATGGTACCCGTAGTCGTTGGAACGGTCATAGCCGTATCCGTTACAATAGGCTGGATGCTTATGATGACGCAATTCTTGTGGAGCTTTGGCACGGCTATTCTTTGTACGGTCCAAGGTATATATTTTGCTGAAATAGCGGATAAAGACATTAGAGGCACCCTCTCGTTGTCAACAAGATACATGTTTAATCTTGGGAATCTTCTAGTCATCGGTGCTGGCCCATTCATAAGTTATCAAACACTAAACTTGTGTTTGCTTTTTATACCGTTGACTTTTTTGATTGCCTCGTTCTGGATACCGGAGACACCATATTATTATCTAAAAGAGGACAAAGTAAACGAGGCCAGGAAGGTTTTGATGCAGCTGCGAGGAATCAAAGATGAAAAGGAATTAGATTCTCAATTAGCATTGATTGAGATGAACGTGAAAGAGGACATGACTAGATCCAGTTCAGTGAAACAGCTCTTCTCGGGAGCCAAATACAGGAAGGCTATTATGATCGTTTCAGGCCTGAAAATTACGTCAATATTTACGGGAACGGTGACAATAAGACAATATTTCGGCAGGATACTACAAGAAGGTCGCATACCCAAAGAAATGGAAGGAACAGTATTTATTGTATTCGGAGCGGTTGGATTCTTGACGGCAATTGTAGCTTCAGTGCTAGTTGATAGATTGGGTCGCCGGCCCCTTCTAATCGGTTCATATATCGTAACCGGTGTATGTCTCGCAGCTATTGGGATTTACTTCTTTCTTCAAGATGCTGTTCTTGTTGATAGCGCAGCCATGTCGCCATATGGGTACTTGCCTTTCATTGGCATACTTCTCGTCACTGTCAGCTCAACGATTGGCTTCAATTCAATAGTGAACTTCATGCCCGCTGAATTATTTCCTTTGAACGTTAAAGCCATTGCTATGTCCCTAAACGGGATATTGAGCGGTGTATTGGGCTTCGCGATTGCCAAATCTTACCAAGATATCAAGGATTTGTGTGGAATTTATACGGTTTTTTGGATTTTCACGGTATTCAGTTGCGCTGGAGGCGTTTTCTGTTATTATTTAGTACCGGAAACTAAGGGTAAGGGCTTAGCGGAAATTCAAGAAATGTTGCAAAGTCCTTGGCTAGAGTTAAGGACGTACGATGGTACCGCGAAGTTTAATATTGGCGATAATAAGGATAGTGAAGAGGAAAGAGCGTCATTAACGGAAGTGACCAAAATTTGA
Protein
MESNRKVQYLAAFSVSLAFLAVGACSAWPTPVLLKFHNNETSVVISNTEISWMLGLNPIGFLIGSFTARFVIDKFGRRYTLLASMVPVVVGTVIAVSVTIGWMLMMTQFLWSFGTAILCTVQGIYFAEIADKDIRGTLSLSTRYMFNLGNLLVIGAGPFISYQTLNLCLLFIPLTFLIASFWIPETPYYYLKEDKVNEARKVLMQLRGIKDEKELDSQLALIEMNVKEDMTRSSSVKQLFSGAKYRKAIMIVSGLKITSIFTGTVTIRQYFGRILQEGRIPKEMEGTVFIVFGAVGFLTAIVASVLVDRLGRRPLLIGSYIVTGVCLAAIGIYFFLQDAVLVDSAAMSPYGYLPFIGILLVTVSSTIGFNSIVNFMPAELFPLNVKAIAMSLNGILSGVLGFAIAKSYQDIKDLCGIYTVFWIFTVFSCAGGVFCYYLVPETKGKGLAEIQEMLQSPWLELRTYDGTAKFNIGDNKDSEEERASLTEVTKI
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9J7W2
A0A212F503
A0A2A4JTV9
A0A3S2L2Y7
A0A2H1WSH1
A0A2H1VJZ6
+ More
A0A2H1WNQ9 A0A2A4JT90 A0A0L7L6Z1 A0A0L7LK83 A0A194QXD6 A0A194QGX6 A0A2A4JTX0 A0A212F501 A0A2H1VF83 A0A154PQ64 A0A1Y1L2V6 A0A3L8DJN2 E9J2F8 A0A0L7R4F0 A0A212ERN0 A0A2A3E771 E2ALN5 A0A0M9A2I9 A0A1B6LDV4 E2BEY2 A0A067R7D8 A0A087ZNC7 A0A1B6H191 F4WJH1 E2A9Z9 A0A151X1H7 A0A2A4JUE8 A0A3S2NTN5 A0A151JVX5 H9IZF0 A0A310SR72 A0A1B6C7Q8 A0A1B6IR35 A0A151IBX7 A0A151I1W8 A0A158NMR6 A0A1B0CUK2 A0A0A8J7X6 A0A2J7RK51 A0A1Y1MME1 A0A194PW18 A0A1S3DL63 A0A2R7X3C9 A0A067RCA0 A0A1S3DQQ7 B0WDB0 A0A1Y1MKJ2 A0A1B6DV63 T1HSD1 A0A0C9R6D5 A0A182NLK4 S4P7H5 A0A069DZ02 A0A084W3Q1 A0A1B6MB27 A0A1Y1MHD8 A0A1B6CWY0 A0A224XH10 E9IVK1 A0A182IRD4 A0A1B6F2Y7 A0A182KRB6 A0A182FF45 W5JRP7 A0A182X3Q8 A0A2H1VY21 R4FM44 Q7QJF0 A0A0C9RC65 A0A182S4P1 E2ADH2 Q16N91 A0A0C9Q8V7 A0A1B6CEQ2 T1DG93 A0A0K8T923 A0A182YRA3 A0A1B6M9Z3 A0A182LZZ1 E0W0A9 A0A182RLR4 A0A0A9WMT7 A0A182I3B1 A0A146LWC6 A0A224XPU4 E2ADH1 A0A084WNA9 A0A182VHI4 A0A182QXE9
A0A2H1WNQ9 A0A2A4JT90 A0A0L7L6Z1 A0A0L7LK83 A0A194QXD6 A0A194QGX6 A0A2A4JTX0 A0A212F501 A0A2H1VF83 A0A154PQ64 A0A1Y1L2V6 A0A3L8DJN2 E9J2F8 A0A0L7R4F0 A0A212ERN0 A0A2A3E771 E2ALN5 A0A0M9A2I9 A0A1B6LDV4 E2BEY2 A0A067R7D8 A0A087ZNC7 A0A1B6H191 F4WJH1 E2A9Z9 A0A151X1H7 A0A2A4JUE8 A0A3S2NTN5 A0A151JVX5 H9IZF0 A0A310SR72 A0A1B6C7Q8 A0A1B6IR35 A0A151IBX7 A0A151I1W8 A0A158NMR6 A0A1B0CUK2 A0A0A8J7X6 A0A2J7RK51 A0A1Y1MME1 A0A194PW18 A0A1S3DL63 A0A2R7X3C9 A0A067RCA0 A0A1S3DQQ7 B0WDB0 A0A1Y1MKJ2 A0A1B6DV63 T1HSD1 A0A0C9R6D5 A0A182NLK4 S4P7H5 A0A069DZ02 A0A084W3Q1 A0A1B6MB27 A0A1Y1MHD8 A0A1B6CWY0 A0A224XH10 E9IVK1 A0A182IRD4 A0A1B6F2Y7 A0A182KRB6 A0A182FF45 W5JRP7 A0A182X3Q8 A0A2H1VY21 R4FM44 Q7QJF0 A0A0C9RC65 A0A182S4P1 E2ADH2 Q16N91 A0A0C9Q8V7 A0A1B6CEQ2 T1DG93 A0A0K8T923 A0A182YRA3 A0A1B6M9Z3 A0A182LZZ1 E0W0A9 A0A182RLR4 A0A0A9WMT7 A0A182I3B1 A0A146LWC6 A0A224XPU4 E2ADH1 A0A084WNA9 A0A182VHI4 A0A182QXE9
Pubmed
EMBL
BABH01020034
AGBW02010278
OWR48815.1
NWSH01000679
PCG74840.1
RSAL01000200
+ More
RVE44474.1 ODYU01010683 SOQ55957.1 ODYU01002974 SOQ41148.1 ODYU01009924 SOQ54668.1 PCG74834.1 JTDY01002524 KOB71243.1 JTDY01000761 KOB75973.1 KQ461155 KPJ08236.1 KQ459232 KPJ02696.1 PCG74842.1 OWR48817.1 ODYU01002251 SOQ39487.1 KQ435031 KZC14029.1 GEZM01066384 GEZM01066383 JAV67914.1 QOIP01000007 RLU20546.1 GL767845 EFZ13014.1 KQ414657 KOC65760.1 AGBW02012985 OWR44135.1 KZ288357 PBC27132.1 GL440609 EFN65644.1 KQ435789 KOX74499.1 GEBQ01018101 JAT21876.1 GL447885 EFN85747.1 KK852657 KDR19246.1 GECZ01001325 JAS68444.1 GL888182 EGI65749.1 GL437987 EFN69767.1 KQ982588 KYQ54104.1 NWSH01000598 PCG75429.1 RSAL01000086 RVE48282.1 KQ981689 KYN37896.1 BABH01014157 KQ760761 OAD59140.1 GEDC01027797 JAS09501.1 GECU01018312 JAS89394.1 KQ978078 KYM97199.1 KQ976593 KYM79675.1 ADTU01020710 AJWK01029294 AB901052 BAQ02352.1 NEVH01002981 PNF41213.1 GEZM01033162 JAV84387.1 KQ459589 KPI97522.1 KK856708 PTY26307.1 KK852731 KDR17493.1 DS231895 EDS44344.1 GEZM01033163 JAV84386.1 GEDC01007769 JAS29529.1 ACPB03022320 GBYB01003549 GBYB01003550 JAG73316.1 JAG73317.1 GAIX01004609 JAA87951.1 GBGD01001495 JAC87394.1 ATLV01020086 KE525294 KFB44845.1 GEBQ01006842 JAT33135.1 GEZM01033161 JAV84388.1 GEDC01019312 JAS17986.1 GFTR01006112 JAW10314.1 GL766258 EFZ15412.1 GECZ01025224 JAS44545.1 ADMH02000549 ETN65958.1 ODYU01004836 SOQ45124.1 GAHY01001869 JAA75641.1 AAAB01008807 EAA04637.4 GBYB01010702 JAG80469.1 GL438750 EFN68534.1 CH477833 EAT35815.1 GBYB01010703 JAG80470.1 GEDC01025390 JAS11908.1 GAMD01002846 JAA98744.1 GBRD01003807 JAG62014.1 GEBQ01007236 JAT32741.1 AXCM01019063 DS235858 EEB19065.1 GBHO01037183 GBHO01034863 JAG06421.1 JAG08741.1 APCN01000204 GDHC01006628 JAQ12001.1 GFTR01006407 JAW10019.1 EFN68533.1 ATLV01024580 KE525352 KFB51703.1 AXCN02002516
RVE44474.1 ODYU01010683 SOQ55957.1 ODYU01002974 SOQ41148.1 ODYU01009924 SOQ54668.1 PCG74834.1 JTDY01002524 KOB71243.1 JTDY01000761 KOB75973.1 KQ461155 KPJ08236.1 KQ459232 KPJ02696.1 PCG74842.1 OWR48817.1 ODYU01002251 SOQ39487.1 KQ435031 KZC14029.1 GEZM01066384 GEZM01066383 JAV67914.1 QOIP01000007 RLU20546.1 GL767845 EFZ13014.1 KQ414657 KOC65760.1 AGBW02012985 OWR44135.1 KZ288357 PBC27132.1 GL440609 EFN65644.1 KQ435789 KOX74499.1 GEBQ01018101 JAT21876.1 GL447885 EFN85747.1 KK852657 KDR19246.1 GECZ01001325 JAS68444.1 GL888182 EGI65749.1 GL437987 EFN69767.1 KQ982588 KYQ54104.1 NWSH01000598 PCG75429.1 RSAL01000086 RVE48282.1 KQ981689 KYN37896.1 BABH01014157 KQ760761 OAD59140.1 GEDC01027797 JAS09501.1 GECU01018312 JAS89394.1 KQ978078 KYM97199.1 KQ976593 KYM79675.1 ADTU01020710 AJWK01029294 AB901052 BAQ02352.1 NEVH01002981 PNF41213.1 GEZM01033162 JAV84387.1 KQ459589 KPI97522.1 KK856708 PTY26307.1 KK852731 KDR17493.1 DS231895 EDS44344.1 GEZM01033163 JAV84386.1 GEDC01007769 JAS29529.1 ACPB03022320 GBYB01003549 GBYB01003550 JAG73316.1 JAG73317.1 GAIX01004609 JAA87951.1 GBGD01001495 JAC87394.1 ATLV01020086 KE525294 KFB44845.1 GEBQ01006842 JAT33135.1 GEZM01033161 JAV84388.1 GEDC01019312 JAS17986.1 GFTR01006112 JAW10314.1 GL766258 EFZ15412.1 GECZ01025224 JAS44545.1 ADMH02000549 ETN65958.1 ODYU01004836 SOQ45124.1 GAHY01001869 JAA75641.1 AAAB01008807 EAA04637.4 GBYB01010702 JAG80469.1 GL438750 EFN68534.1 CH477833 EAT35815.1 GBYB01010703 JAG80470.1 GEDC01025390 JAS11908.1 GAMD01002846 JAA98744.1 GBRD01003807 JAG62014.1 GEBQ01007236 JAT32741.1 AXCM01019063 DS235858 EEB19065.1 GBHO01037183 GBHO01034863 JAG06421.1 JAG08741.1 APCN01000204 GDHC01006628 JAQ12001.1 GFTR01006407 JAW10019.1 EFN68533.1 ATLV01024580 KE525352 KFB51703.1 AXCN02002516
Proteomes
UP000005204
UP000007151
UP000218220
UP000283053
UP000037510
UP000053240
+ More
UP000053268 UP000076502 UP000279307 UP000053825 UP000242457 UP000000311 UP000053105 UP000008237 UP000027135 UP000005203 UP000007755 UP000075809 UP000078541 UP000078542 UP000078540 UP000005205 UP000092461 UP000235965 UP000079169 UP000002320 UP000015103 UP000075884 UP000030765 UP000075880 UP000075882 UP000069272 UP000000673 UP000007062 UP000075900 UP000008820 UP000076408 UP000075883 UP000009046 UP000075840 UP000075886
UP000053268 UP000076502 UP000279307 UP000053825 UP000242457 UP000000311 UP000053105 UP000008237 UP000027135 UP000005203 UP000007755 UP000075809 UP000078541 UP000078542 UP000078540 UP000005205 UP000092461 UP000235965 UP000079169 UP000002320 UP000015103 UP000075884 UP000030765 UP000075880 UP000075882 UP000069272 UP000000673 UP000007062 UP000075900 UP000008820 UP000076408 UP000075883 UP000009046 UP000075840 UP000075886
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9J7W2
A0A212F503
A0A2A4JTV9
A0A3S2L2Y7
A0A2H1WSH1
A0A2H1VJZ6
+ More
A0A2H1WNQ9 A0A2A4JT90 A0A0L7L6Z1 A0A0L7LK83 A0A194QXD6 A0A194QGX6 A0A2A4JTX0 A0A212F501 A0A2H1VF83 A0A154PQ64 A0A1Y1L2V6 A0A3L8DJN2 E9J2F8 A0A0L7R4F0 A0A212ERN0 A0A2A3E771 E2ALN5 A0A0M9A2I9 A0A1B6LDV4 E2BEY2 A0A067R7D8 A0A087ZNC7 A0A1B6H191 F4WJH1 E2A9Z9 A0A151X1H7 A0A2A4JUE8 A0A3S2NTN5 A0A151JVX5 H9IZF0 A0A310SR72 A0A1B6C7Q8 A0A1B6IR35 A0A151IBX7 A0A151I1W8 A0A158NMR6 A0A1B0CUK2 A0A0A8J7X6 A0A2J7RK51 A0A1Y1MME1 A0A194PW18 A0A1S3DL63 A0A2R7X3C9 A0A067RCA0 A0A1S3DQQ7 B0WDB0 A0A1Y1MKJ2 A0A1B6DV63 T1HSD1 A0A0C9R6D5 A0A182NLK4 S4P7H5 A0A069DZ02 A0A084W3Q1 A0A1B6MB27 A0A1Y1MHD8 A0A1B6CWY0 A0A224XH10 E9IVK1 A0A182IRD4 A0A1B6F2Y7 A0A182KRB6 A0A182FF45 W5JRP7 A0A182X3Q8 A0A2H1VY21 R4FM44 Q7QJF0 A0A0C9RC65 A0A182S4P1 E2ADH2 Q16N91 A0A0C9Q8V7 A0A1B6CEQ2 T1DG93 A0A0K8T923 A0A182YRA3 A0A1B6M9Z3 A0A182LZZ1 E0W0A9 A0A182RLR4 A0A0A9WMT7 A0A182I3B1 A0A146LWC6 A0A224XPU4 E2ADH1 A0A084WNA9 A0A182VHI4 A0A182QXE9
A0A2H1WNQ9 A0A2A4JT90 A0A0L7L6Z1 A0A0L7LK83 A0A194QXD6 A0A194QGX6 A0A2A4JTX0 A0A212F501 A0A2H1VF83 A0A154PQ64 A0A1Y1L2V6 A0A3L8DJN2 E9J2F8 A0A0L7R4F0 A0A212ERN0 A0A2A3E771 E2ALN5 A0A0M9A2I9 A0A1B6LDV4 E2BEY2 A0A067R7D8 A0A087ZNC7 A0A1B6H191 F4WJH1 E2A9Z9 A0A151X1H7 A0A2A4JUE8 A0A3S2NTN5 A0A151JVX5 H9IZF0 A0A310SR72 A0A1B6C7Q8 A0A1B6IR35 A0A151IBX7 A0A151I1W8 A0A158NMR6 A0A1B0CUK2 A0A0A8J7X6 A0A2J7RK51 A0A1Y1MME1 A0A194PW18 A0A1S3DL63 A0A2R7X3C9 A0A067RCA0 A0A1S3DQQ7 B0WDB0 A0A1Y1MKJ2 A0A1B6DV63 T1HSD1 A0A0C9R6D5 A0A182NLK4 S4P7H5 A0A069DZ02 A0A084W3Q1 A0A1B6MB27 A0A1Y1MHD8 A0A1B6CWY0 A0A224XH10 E9IVK1 A0A182IRD4 A0A1B6F2Y7 A0A182KRB6 A0A182FF45 W5JRP7 A0A182X3Q8 A0A2H1VY21 R4FM44 Q7QJF0 A0A0C9RC65 A0A182S4P1 E2ADH2 Q16N91 A0A0C9Q8V7 A0A1B6CEQ2 T1DG93 A0A0K8T923 A0A182YRA3 A0A1B6M9Z3 A0A182LZZ1 E0W0A9 A0A182RLR4 A0A0A9WMT7 A0A182I3B1 A0A146LWC6 A0A224XPU4 E2ADH1 A0A084WNA9 A0A182VHI4 A0A182QXE9
PDB
5EQI
E-value=8.96737e-22,
Score=257
Ontologies
GO
Topology
SignalP
Position: 1 - 27,
Likelihood: 0.765891
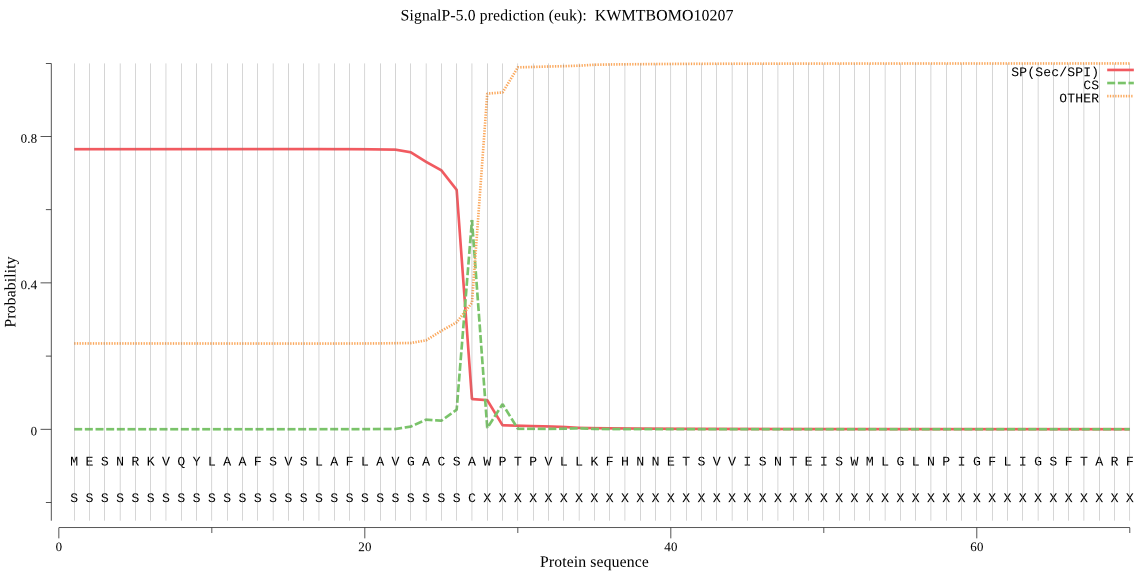
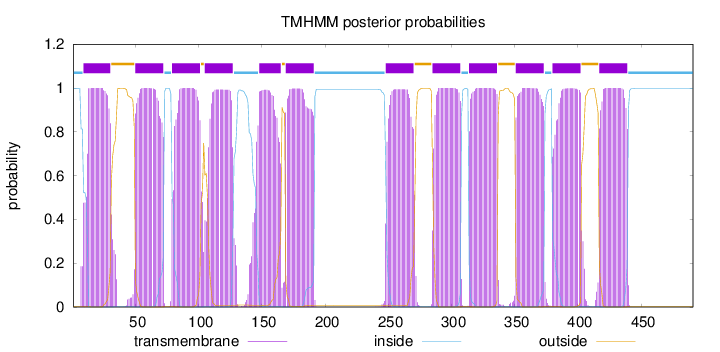
Length:
491
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
264.99252
Exp number, first 60 AAs:
32.67297
Total prob of N-in:
0.99867
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 30
outside
31 - 49
TMhelix
50 - 72
inside
73 - 78
TMhelix
79 - 101
outside
102 - 104
TMhelix
105 - 127
inside
128 - 147
TMhelix
148 - 165
outside
166 - 168
TMhelix
169 - 191
inside
192 - 247
TMhelix
248 - 270
outside
271 - 284
TMhelix
285 - 307
inside
308 - 313
TMhelix
314 - 336
outside
337 - 350
TMhelix
351 - 373
inside
374 - 379
TMhelix
380 - 402
outside
403 - 416
TMhelix
417 - 439
inside
440 - 491
Population Genetic Test Statistics
Pi
170.176444
Theta
165.169887
Tajima's D
0.329551
CLR
0.293237
CSRT
0.468576571171441
Interpretation
Uncertain
