Gene
KWMTBOMO10206
Pre Gene Modal
BGIBMGA005605
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.836
Sequence
CDS
ATGGCTGTTGGAACGATTATATTACAAATCGCTACTAAAGCTTGGATGCTTTACATGACTAGGCTCATTTGGGGATTTTCAACGGGAATGTTTATGACGGTGTCCTTTATATACTTCGTAGAGATAGCCGACAAAGAGATAAGAGGGAAACTATCTGTAGCTATTCGATTCATGTTTAACTTCGGCAGTTTCCTCGTTGTATCTGTGGGCTCTTTCGTCTCATACACCACGTTGAACAGAATGCTGATAGTATTGCCTATAGGTCACTTTGTGGCCTGTTGGTTTATACCGGAAACTCCGCATTTCTACATAAAGGAGGGTAAAGTTGAAGCCGCTGAGAAATCGCTGCAGTTGCTGCACGGATGTAAGGATCAAAATGAATTACAGAGCCGTCTTGAGTCACTTCGATCGGACGTGAAGAAGCAAACGCGTCGTCCGGGCACATTTAAAGAATTGTTTACTGGCAAACAATACAGACGAGCTATAGTCATTGTCATTGGTATTAAAACGACACAGATCATGTCTGGGTCGCTTCCCATCCAGCAATACGCCGGTCTTATTATCCAAGAGAGTCACGCGGATATTCCTCTATCTGCAGCTTTGATTAGCTTCGGTGCTGTCAGATTTGTCGCAGGTATAATTTCATCGTATCTAGTCGATAAAGTAGGGCGTCGCCCTCTGCTGATAGTTACATATATCTGCACAGGCGTGACTCTAATCATCGTCGGAATTTACTTTTACTTTCAAGAAGTGGTCGGCATTGATATCGAAACTTCTAGTCCTTTGAGATACATTCCATTTGTAGGTGTATTCCTCAGTATTAGTTTGTCCACAATCGGCTATGACTCTTTGGTGTACATAATTCCGGCTGAAATATTTCCGATTAACGTCAAATCTCTAGCTATTACTTATTTGAATATATATGGCGGTCTAATGAATTTCTTAGCTGTGAAAGGATTTCAGAATATGAGGGACTGGACGGGTCTTTATGGCGTGTTCTGGTATTTTGCGGCATTTGCTCTGTTAGGCGGGACGTTCTCGTTTTTTATGGCGCCAGAAACAAAAGGTAAAAGCCTCAGGGAGATATTGATTGAATTGCAAGGAGACTTATACGAGGAACCAGAGGAAAGAAAATTAAACGGTACTAAGGGTGATGTTCAAAATAGGGGCGTCAAAGAAAGTTGTGAATTAACAGAATTAGCGTGTAAAAATGAAGAGAACAATAGGAATAGTACTGGAGATTCGTGA
Protein
MAVGTIILQIATKAWMLYMTRLIWGFSTGMFMTVSFIYFVEIADKEIRGKLSVAIRFMFNFGSFLVVSVGSFVSYTTLNRMLIVLPIGHFVACWFIPETPHFYIKEGKVEAAEKSLQLLHGCKDQNELQSRLESLRSDVKKQTRRPGTFKELFTGKQYRRAIVIVIGIKTTQIMSGSLPIQQYAGLIIQESHADIPLSAALISFGAVRFVAGIISSYLVDKVGRRPLLIVTYICTGVTLIIVGIYFYFQEVVGIDIETSSPLRYIPFVGVFLSISLSTIGYDSLVYIIPAEIFPINVKSLAITYLNIYGGLMNFLAVKGFQNMRDWTGLYGVFWYFAAFALLGGTFSFFMAPETKGKSLREILIELQGDLYEEPEERKLNGTKGDVQNRGVKESCELTELACKNEENNRNSTGDS
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9J7W3
A0A2A4JT90
A0A2H1VJZ6
A0A2H1WNQ9
A0A0L7LK83
A0A0L7L6Z1
+ More
A0A2A4JTV9 A0A3S2L2Y7 A0A2H1WSH1 A0A212F503 A0A194QAX5 H9J7W2 A0A0L7L6Z3 A0A2A4JTX0 A0A0L7K3V1 A0A3S2LI48 A0A2H1VF83 A0A194QXD6 A0A194QGX6 A0A0L7LKT1 A0A212F501 A0A1Y1L2V6 N6TV36 U4UVU4 A0A3L8DJN2 A0A2A4JUE8 A0A154PQ64 W4VS78 A0A2H1VY21 A0A1W7R5M1 A0A194R1W3 A0A087ZNC7 A0A212ERN0 S4P7H5 H9IZE9 A0A194PW18 A0A1Y1MME1 E2ALN5 A0A084WNA8 A0A182V7I7 A0A182JUH6 Q16N91 A0A232ERM9 A0A182I3B0 E2BEY2 Q7QJE9 A0A182J5I6 A0A1Y1MKJ2 A0A151X1H7 A0A212ERK6 A0A3S2NTN5 A0A182X3Q9 A0A182TN07 A0A2A3E771 F4WJH1 A0A182KRC0 A0A182FF45 A0A1S4F5X0 Q16ZD3 Q17EH6 A0A1Y1LTF4 W5JRP7 A0A195DBN7 A0A182RSN7 A0A1Y1MHD8 A0A151IBX7 A0A1Q3FJP9 A0A182PQF0 A0A151JVX5 A0A182XYW5 A0A182JJ49 A0A2M4BL39 A0A3S2LAH9 A0A2M4CKN1 A0A1B6H191 A0A2P8YZ38 E9J2F8 A0A1Q3FJI4 A0A182NHV3 K7IWX4 A0A1Y1NBK4 B0X8T3 A0A1B6IR35 A0A2M4CKL9 A0A0L7LKK9 A0A3S2NG77 A0A182S6W7 A0A023ET97 A0A2H1VF40 A0A023EU90 A0A2W1BU14 A0A2A4JPP7 A0A182GCG2 D6WET6 A0A0L7R4F0 A0A1B0CUK2 U4UPG3 A0A182MUN7 A0A182WJ81
A0A2A4JTV9 A0A3S2L2Y7 A0A2H1WSH1 A0A212F503 A0A194QAX5 H9J7W2 A0A0L7L6Z3 A0A2A4JTX0 A0A0L7K3V1 A0A3S2LI48 A0A2H1VF83 A0A194QXD6 A0A194QGX6 A0A0L7LKT1 A0A212F501 A0A1Y1L2V6 N6TV36 U4UVU4 A0A3L8DJN2 A0A2A4JUE8 A0A154PQ64 W4VS78 A0A2H1VY21 A0A1W7R5M1 A0A194R1W3 A0A087ZNC7 A0A212ERN0 S4P7H5 H9IZE9 A0A194PW18 A0A1Y1MME1 E2ALN5 A0A084WNA8 A0A182V7I7 A0A182JUH6 Q16N91 A0A232ERM9 A0A182I3B0 E2BEY2 Q7QJE9 A0A182J5I6 A0A1Y1MKJ2 A0A151X1H7 A0A212ERK6 A0A3S2NTN5 A0A182X3Q9 A0A182TN07 A0A2A3E771 F4WJH1 A0A182KRC0 A0A182FF45 A0A1S4F5X0 Q16ZD3 Q17EH6 A0A1Y1LTF4 W5JRP7 A0A195DBN7 A0A182RSN7 A0A1Y1MHD8 A0A151IBX7 A0A1Q3FJP9 A0A182PQF0 A0A151JVX5 A0A182XYW5 A0A182JJ49 A0A2M4BL39 A0A3S2LAH9 A0A2M4CKN1 A0A1B6H191 A0A2P8YZ38 E9J2F8 A0A1Q3FJI4 A0A182NHV3 K7IWX4 A0A1Y1NBK4 B0X8T3 A0A1B6IR35 A0A2M4CKL9 A0A0L7LKK9 A0A3S2NG77 A0A182S6W7 A0A023ET97 A0A2H1VF40 A0A023EU90 A0A2W1BU14 A0A2A4JPP7 A0A182GCG2 D6WET6 A0A0L7R4F0 A0A1B0CUK2 U4UPG3 A0A182MUN7 A0A182WJ81
Pubmed
EMBL
BABH01020035
NWSH01000679
PCG74834.1
ODYU01002974
SOQ41148.1
ODYU01009924
+ More
SOQ54668.1 JTDY01000761 KOB75973.1 JTDY01002524 KOB71243.1 PCG74840.1 RSAL01000200 RVE44474.1 ODYU01010683 SOQ55957.1 AGBW02010278 OWR48815.1 KQ459232 KPJ02693.1 BABH01020034 KOB71242.1 PCG74842.1 JTDY01011397 KOB56606.1 RSAL01000110 RVE47159.1 ODYU01002251 SOQ39487.1 KQ461155 KPJ08236.1 KPJ02696.1 KOB75974.1 OWR48817.1 GEZM01066384 GEZM01066383 JAV67914.1 APGK01053692 APGK01053693 KB741231 ENN72261.1 KB632387 ERL94361.1 QOIP01000007 RLU20546.1 NWSH01000598 PCG75429.1 KQ435031 KZC14029.1 GAPU01000012 JAB84606.1 ODYU01004836 SOQ45124.1 GEHC01001176 JAV46469.1 KQ460870 KPJ11682.1 AGBW02012985 OWR44135.1 GAIX01004609 JAA87951.1 BABH01014162 KQ459589 KPI97522.1 GEZM01033162 JAV84387.1 GL440609 EFN65644.1 ATLV01024580 KE525352 KFB51702.1 CH477833 EAT35815.1 NNAY01002587 OXU20982.1 APCN01000204 GL447885 EFN85747.1 AAAB01008807 EAA04375.4 GEZM01033163 JAV84386.1 KQ982588 KYQ54104.1 OWR44136.1 RSAL01000086 RVE48282.1 KZ288357 PBC27132.1 GL888182 EGI65749.1 CH477492 EAT40014.1 CH477282 EAT44903.1 GEZM01050479 JAV75580.1 ADMH02000549 ETN65958.1 KQ981010 KYN10335.1 GEZM01033161 JAV84388.1 KQ978078 KYM97199.1 GFDL01007246 JAV27799.1 KQ981689 KYN37896.1 GGFJ01004530 MBW53671.1 RSAL01000321 RVE42560.1 GGFL01001699 MBW65877.1 GECZ01001325 JAS68444.1 PYGN01000276 PSN49516.1 GL767845 EFZ13014.1 GFDL01007265 JAV27780.1 AAZX01006527 AAZX01008308 GEZM01007168 GEZM01007167 JAV95324.1 DS232501 EDS42675.1 GECU01018312 JAS89394.1 GGFL01001698 MBW65876.1 JTDY01000837 KOB75721.1 RSAL01000118 RVE46763.1 GAPW01001080 JAC12518.1 ODYU01001965 SOQ38884.1 GAPW01001122 JAC12476.1 KZ149936 PZC77145.1 NWSH01000916 PCG73554.1 JXUM01054970 JXUM01054971 KQ561845 KXJ77365.1 KQ971318 EFA00434.1 KQ414657 KOC65760.1 AJWK01029294 ERL94363.1 AXCM01011849
SOQ54668.1 JTDY01000761 KOB75973.1 JTDY01002524 KOB71243.1 PCG74840.1 RSAL01000200 RVE44474.1 ODYU01010683 SOQ55957.1 AGBW02010278 OWR48815.1 KQ459232 KPJ02693.1 BABH01020034 KOB71242.1 PCG74842.1 JTDY01011397 KOB56606.1 RSAL01000110 RVE47159.1 ODYU01002251 SOQ39487.1 KQ461155 KPJ08236.1 KPJ02696.1 KOB75974.1 OWR48817.1 GEZM01066384 GEZM01066383 JAV67914.1 APGK01053692 APGK01053693 KB741231 ENN72261.1 KB632387 ERL94361.1 QOIP01000007 RLU20546.1 NWSH01000598 PCG75429.1 KQ435031 KZC14029.1 GAPU01000012 JAB84606.1 ODYU01004836 SOQ45124.1 GEHC01001176 JAV46469.1 KQ460870 KPJ11682.1 AGBW02012985 OWR44135.1 GAIX01004609 JAA87951.1 BABH01014162 KQ459589 KPI97522.1 GEZM01033162 JAV84387.1 GL440609 EFN65644.1 ATLV01024580 KE525352 KFB51702.1 CH477833 EAT35815.1 NNAY01002587 OXU20982.1 APCN01000204 GL447885 EFN85747.1 AAAB01008807 EAA04375.4 GEZM01033163 JAV84386.1 KQ982588 KYQ54104.1 OWR44136.1 RSAL01000086 RVE48282.1 KZ288357 PBC27132.1 GL888182 EGI65749.1 CH477492 EAT40014.1 CH477282 EAT44903.1 GEZM01050479 JAV75580.1 ADMH02000549 ETN65958.1 KQ981010 KYN10335.1 GEZM01033161 JAV84388.1 KQ978078 KYM97199.1 GFDL01007246 JAV27799.1 KQ981689 KYN37896.1 GGFJ01004530 MBW53671.1 RSAL01000321 RVE42560.1 GGFL01001699 MBW65877.1 GECZ01001325 JAS68444.1 PYGN01000276 PSN49516.1 GL767845 EFZ13014.1 GFDL01007265 JAV27780.1 AAZX01006527 AAZX01008308 GEZM01007168 GEZM01007167 JAV95324.1 DS232501 EDS42675.1 GECU01018312 JAS89394.1 GGFL01001698 MBW65876.1 JTDY01000837 KOB75721.1 RSAL01000118 RVE46763.1 GAPW01001080 JAC12518.1 ODYU01001965 SOQ38884.1 GAPW01001122 JAC12476.1 KZ149936 PZC77145.1 NWSH01000916 PCG73554.1 JXUM01054970 JXUM01054971 KQ561845 KXJ77365.1 KQ971318 EFA00434.1 KQ414657 KOC65760.1 AJWK01029294 ERL94363.1 AXCM01011849
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000007151
UP000053268
+ More
UP000053240 UP000019118 UP000030742 UP000279307 UP000076502 UP000005203 UP000000311 UP000030765 UP000075903 UP000075881 UP000008820 UP000215335 UP000075840 UP000008237 UP000007062 UP000075880 UP000075809 UP000076407 UP000075902 UP000242457 UP000007755 UP000075882 UP000069272 UP000000673 UP000078492 UP000075900 UP000078542 UP000075885 UP000078541 UP000076408 UP000245037 UP000075884 UP000002358 UP000002320 UP000075901 UP000069940 UP000249989 UP000007266 UP000053825 UP000092461 UP000075883 UP000075920
UP000053240 UP000019118 UP000030742 UP000279307 UP000076502 UP000005203 UP000000311 UP000030765 UP000075903 UP000075881 UP000008820 UP000215335 UP000075840 UP000008237 UP000007062 UP000075880 UP000075809 UP000076407 UP000075902 UP000242457 UP000007755 UP000075882 UP000069272 UP000000673 UP000078492 UP000075900 UP000078542 UP000075885 UP000078541 UP000076408 UP000245037 UP000075884 UP000002358 UP000002320 UP000075901 UP000069940 UP000249989 UP000007266 UP000053825 UP000092461 UP000075883 UP000075920
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9J7W3
A0A2A4JT90
A0A2H1VJZ6
A0A2H1WNQ9
A0A0L7LK83
A0A0L7L6Z1
+ More
A0A2A4JTV9 A0A3S2L2Y7 A0A2H1WSH1 A0A212F503 A0A194QAX5 H9J7W2 A0A0L7L6Z3 A0A2A4JTX0 A0A0L7K3V1 A0A3S2LI48 A0A2H1VF83 A0A194QXD6 A0A194QGX6 A0A0L7LKT1 A0A212F501 A0A1Y1L2V6 N6TV36 U4UVU4 A0A3L8DJN2 A0A2A4JUE8 A0A154PQ64 W4VS78 A0A2H1VY21 A0A1W7R5M1 A0A194R1W3 A0A087ZNC7 A0A212ERN0 S4P7H5 H9IZE9 A0A194PW18 A0A1Y1MME1 E2ALN5 A0A084WNA8 A0A182V7I7 A0A182JUH6 Q16N91 A0A232ERM9 A0A182I3B0 E2BEY2 Q7QJE9 A0A182J5I6 A0A1Y1MKJ2 A0A151X1H7 A0A212ERK6 A0A3S2NTN5 A0A182X3Q9 A0A182TN07 A0A2A3E771 F4WJH1 A0A182KRC0 A0A182FF45 A0A1S4F5X0 Q16ZD3 Q17EH6 A0A1Y1LTF4 W5JRP7 A0A195DBN7 A0A182RSN7 A0A1Y1MHD8 A0A151IBX7 A0A1Q3FJP9 A0A182PQF0 A0A151JVX5 A0A182XYW5 A0A182JJ49 A0A2M4BL39 A0A3S2LAH9 A0A2M4CKN1 A0A1B6H191 A0A2P8YZ38 E9J2F8 A0A1Q3FJI4 A0A182NHV3 K7IWX4 A0A1Y1NBK4 B0X8T3 A0A1B6IR35 A0A2M4CKL9 A0A0L7LKK9 A0A3S2NG77 A0A182S6W7 A0A023ET97 A0A2H1VF40 A0A023EU90 A0A2W1BU14 A0A2A4JPP7 A0A182GCG2 D6WET6 A0A0L7R4F0 A0A1B0CUK2 U4UPG3 A0A182MUN7 A0A182WJ81
A0A2A4JTV9 A0A3S2L2Y7 A0A2H1WSH1 A0A212F503 A0A194QAX5 H9J7W2 A0A0L7L6Z3 A0A2A4JTX0 A0A0L7K3V1 A0A3S2LI48 A0A2H1VF83 A0A194QXD6 A0A194QGX6 A0A0L7LKT1 A0A212F501 A0A1Y1L2V6 N6TV36 U4UVU4 A0A3L8DJN2 A0A2A4JUE8 A0A154PQ64 W4VS78 A0A2H1VY21 A0A1W7R5M1 A0A194R1W3 A0A087ZNC7 A0A212ERN0 S4P7H5 H9IZE9 A0A194PW18 A0A1Y1MME1 E2ALN5 A0A084WNA8 A0A182V7I7 A0A182JUH6 Q16N91 A0A232ERM9 A0A182I3B0 E2BEY2 Q7QJE9 A0A182J5I6 A0A1Y1MKJ2 A0A151X1H7 A0A212ERK6 A0A3S2NTN5 A0A182X3Q9 A0A182TN07 A0A2A3E771 F4WJH1 A0A182KRC0 A0A182FF45 A0A1S4F5X0 Q16ZD3 Q17EH6 A0A1Y1LTF4 W5JRP7 A0A195DBN7 A0A182RSN7 A0A1Y1MHD8 A0A151IBX7 A0A1Q3FJP9 A0A182PQF0 A0A151JVX5 A0A182XYW5 A0A182JJ49 A0A2M4BL39 A0A3S2LAH9 A0A2M4CKN1 A0A1B6H191 A0A2P8YZ38 E9J2F8 A0A1Q3FJI4 A0A182NHV3 K7IWX4 A0A1Y1NBK4 B0X8T3 A0A1B6IR35 A0A2M4CKL9 A0A0L7LKK9 A0A3S2NG77 A0A182S6W7 A0A023ET97 A0A2H1VF40 A0A023EU90 A0A2W1BU14 A0A2A4JPP7 A0A182GCG2 D6WET6 A0A0L7R4F0 A0A1B0CUK2 U4UPG3 A0A182MUN7 A0A182WJ81
PDB
4ZWC
E-value=3.69082e-17,
Score=216
Ontologies
GO
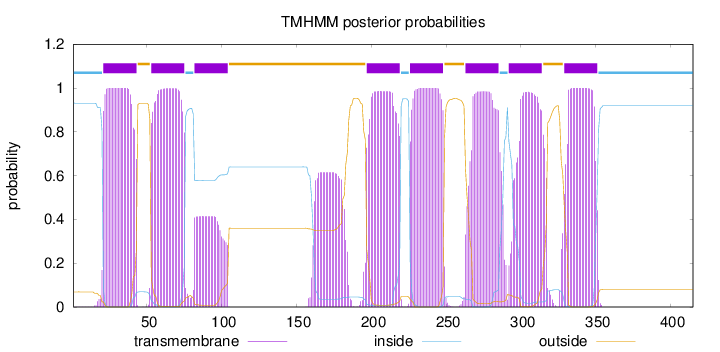
Topology
Length:
415
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
176.74437
Exp number, first 60 AAs:
30.00166
Total prob of N-in:
0.93114
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 52
TMhelix
53 - 75
inside
76 - 81
TMhelix
82 - 104
outside
105 - 196
TMhelix
197 - 219
inside
220 - 225
TMhelix
226 - 248
outside
249 - 262
TMhelix
263 - 285
inside
286 - 291
TMhelix
292 - 314
outside
315 - 328
TMhelix
329 - 351
inside
352 - 415
Population Genetic Test Statistics
Pi
367.194966
Theta
216.956575
Tajima's D
2.109942
CLR
0.079575
CSRT
0.899505024748763
Interpretation
Uncertain
