Gene
KWMTBOMO10203
Annotation
PREDICTED:_uncharacterized_protein_LOC105841331_[Bombyx_mori]
Location in the cell
Peroxisomal Reliability : 0.974
Sequence
CDS
ATGTCTAAAGCCGAACAGGAATGTGAGGATTTTTATGATTTAAATACATACCGCGATAACACAACTGGACGTTACGTCACCGCTTTGCCTTTCTTGAGGTCGCCTACCGCTCTGGGCGATTCTCTTACAACCGCACGTCGCTGTCTGTCGTCACTCGAACGCAGATTTAAGACAAATATCGCTTTAAAAACTGAATACGATGCCGCGATGAGAGACTATCTCGAACGGAACATCATTTCACCCGCACCCGCTCTTACTGATACGAATTATTACGTCATCCCTCACCACGGTATAAGCCGAGAAGACAAACCGCTCCGCATTGTACTGAACGCAACTGCACGATGCATAGATACAAACGTTTCATTGAACGATTGTTTACATTCCGGTCCTAATTTGCAAGCAGACTTAACCGCGATTATTTTAAAATTCAGACTGTTTCCGATTGCTTTAACCGCCGACATCAAACAAATGTTTTTGGCTATAGGCATTCGTGAGGAAGACCGCCGCTTTTTGAATATACTATACAAATTTGACGAAGATGATACTGCGAAAATGTATCATTTTAATCGCGTTTGTTTTGGTCTACGGTGCTCGCCGTACCTTGCTATGAAGACAGTTAAGCGTCTCGCCCGCGATGAAACTGATTTCCCGATCGCTTCAGAAATCGCTAGTGATCATCTTTATATGGATGATCTAGGAGTTTCGGTAGATACGCCCGAGAATGGAATTCAGTTGGCTCATGATTTAATCGAACTATTCAAACGTGGTGGTTTCGACTTAGTTAAATTCGTTAGCAATTCGCCCGAGGTGTTAAAATCGATTCCGCAAGCCAACCGCTTATCTTCAAAAATTGAATTCGACAAGCAGGATAATATGAAGCTCCTAGGTATAATCTGGTCACCGGTCGAGGATGAATTTTCAATCGCTATTAAATTCAACCCGCAAGCAAAATGTACAAAGCGCCTTATGCTTTCCACAATCGCTAAACTGTGGGACATAAACGGATTTTGTGCCCCTGTCATTTTACACGCAAAATTGTTAATAAAAGCTCTCTGGGTAGCTCAGATTGATTGGGATGCAGAGCCCCCAAGCGACATCTGTGCTGCATGGAAACGCTTTCTTAATGAACTCCCCGCTCTGCAAGACTTTAAAATACCTCGCTTCGTAGGCATCGTGCCCGAGTCCCGTGTCATACTACTGGGATTTGCGGACGCTTCTGAAAAAGCGATGGGTGCTTCTGTGTACATTTGTGTACAACCGGCTTCCGGACCAAATGAAGTCAATTTAATTGGTGCCAAAAGCAAGGTCGCGCCGCTCAAGAAAACTCACCGCCTTTACAATGGACAATGGGCATAA
Protein
MSKAEQECEDFYDLNTYRDNTTGRYVTALPFLRSPTALGDSLTTARRCLSSLERRFKTNIALKTEYDAAMRDYLERNIISPAPALTDTNYYVIPHHGISREDKPLRIVLNATARCIDTNVSLNDCLHSGPNLQADLTAIILKFRLFPIALTADIKQMFLAIGIREEDRRFLNILYKFDEDDTAKMYHFNRVCFGLRCSPYLAMKTVKRLARDETDFPIASEIASDHLYMDDLGVSVDTPENGIQLAHDLIELFKRGGFDLVKFVSNSPEVLKSIPQANRLSSKIEFDKQDNMKLLGIIWSPVEDEFSIAIKFNPQAKCTKRLMLSTIAKLWDINGFCAPVILHAKLLIKALWVAQIDWDAEPPSDICAAWKRFLNELPALQDFKIPRFVGIVPESRVILLGFADASEKAMGASVYICVQPASGPNEVNLIGAKSKVAPLKKTHRLYNGQWA
Summary
Uniprot
A0A0A9Z0W7
A0A3Q0J0P0
A0A1S3DMT1
A0A0A9W266
A0A0A9VTB0
A0A023F096
+ More
A0A0A9VUS3 A0A3Q0JFQ3 A0A1Y1LIC8 A0A3Q0JJJ7 A0A1S3DM22 A0A1S3DIZ2 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A1S4EIX5 S4NIN9 A0A0J7N890 A0A1B6LLT3 A0A1B6MT91 A0A1B6KIX6 A0A1S3CUQ4 A0A3Q0IQQ3 A0A3Q0JCL2 A0A1S3DHM0 A0A146M0N4 V5GHM7 A0A1S3DH93 A0A1Y1KMX0 A0A1S3DMK8 A0A1B6GGX5 A0A0J7K7L1 A0A3Q0J7N1 A0A3Q0JBZ9 A0A3Q0J7K9 A0A0J7KB34 J9KD60 A0A0J7K7U0 A0A1S3DIE2 J9JVD6 A0A3Q0JDB5 A0A3Q0JI79 A0A1S3DHZ9 A0A023F0C4 A0A023EYR9 A0A0J7N465 A0A1S3DJJ8 X1WY64 A0A2S2PM34 J9JUM9 A0A1S3DTS8 X1WZY4 A0A1B6MIG8 A0A0A9WHD6 A0A1S3DNV9 A0A3S2TB02 A0A146KRI6 D7EKN0 W8AH72 A0A1Y1LM33 A0A1S3DBK1 A0A3Q0IZW4 A0A1Y1LIG5 A0A1Y1LBZ3 A0A0J7KDN6 X1WT50 A0A3L8DBI7 X1XU06 V5GC07 A0A0A9WWW3 A0A0J7N5Z0 A0A1B6M7K2 A0A1S3DMN9 A0A1B6EKI9 A0A0A9VYL4 A0A2S2NCY5 A0A0A9VSF7 A0A1S3DMW8 X1WVF9 X1X9U0 W8BU98 A0A0A9X8U7 A0A226D417 A0A0A9XDK6 A0A0J7K952 A0A0J7K9I5 J9K0A1 J9JUK4 A0A0A9YU87 A0A2M4AAX1 A0A2S2NNC8 A0A0A9YQQ8 A0A2M4AAS4 A0A2S2N9D0 A0A0A9XBI3 A0A2S2R014 A0A2S2QLL5 J9K351 A0A2M4AAR9 A0A2S2NFZ7
A0A0A9VUS3 A0A3Q0JFQ3 A0A1Y1LIC8 A0A3Q0JJJ7 A0A1S3DM22 A0A1S3DIZ2 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A1S4EIX5 S4NIN9 A0A0J7N890 A0A1B6LLT3 A0A1B6MT91 A0A1B6KIX6 A0A1S3CUQ4 A0A3Q0IQQ3 A0A3Q0JCL2 A0A1S3DHM0 A0A146M0N4 V5GHM7 A0A1S3DH93 A0A1Y1KMX0 A0A1S3DMK8 A0A1B6GGX5 A0A0J7K7L1 A0A3Q0J7N1 A0A3Q0JBZ9 A0A3Q0J7K9 A0A0J7KB34 J9KD60 A0A0J7K7U0 A0A1S3DIE2 J9JVD6 A0A3Q0JDB5 A0A3Q0JI79 A0A1S3DHZ9 A0A023F0C4 A0A023EYR9 A0A0J7N465 A0A1S3DJJ8 X1WY64 A0A2S2PM34 J9JUM9 A0A1S3DTS8 X1WZY4 A0A1B6MIG8 A0A0A9WHD6 A0A1S3DNV9 A0A3S2TB02 A0A146KRI6 D7EKN0 W8AH72 A0A1Y1LM33 A0A1S3DBK1 A0A3Q0IZW4 A0A1Y1LIG5 A0A1Y1LBZ3 A0A0J7KDN6 X1WT50 A0A3L8DBI7 X1XU06 V5GC07 A0A0A9WWW3 A0A0J7N5Z0 A0A1B6M7K2 A0A1S3DMN9 A0A1B6EKI9 A0A0A9VYL4 A0A2S2NCY5 A0A0A9VSF7 A0A1S3DMW8 X1WVF9 X1X9U0 W8BU98 A0A0A9X8U7 A0A226D417 A0A0A9XDK6 A0A0J7K952 A0A0J7K9I5 J9K0A1 J9JUK4 A0A0A9YU87 A0A2M4AAX1 A0A2S2NNC8 A0A0A9YQQ8 A0A2M4AAS4 A0A2S2N9D0 A0A0A9XBI3 A0A2S2R014 A0A2S2QLL5 J9K351 A0A2M4AAR9 A0A2S2NFZ7
EMBL
GBHO01005525
JAG38079.1
GBHO01044644
JAF98959.1
GBHO01044645
JAF98958.1
+ More
GBBI01004276 JAC14436.1 GBHO01044643 JAF98960.1 GEZM01054789 JAV73399.1 GAIX01013989 JAA78571.1 LBMM01008464 KMQ88855.1 GEBQ01015458 JAT24519.1 GEBQ01000825 JAT39152.1 GEBQ01028569 JAT11408.1 GDHC01006469 JAQ12160.1 GALX01004947 JAB63519.1 GEZM01085145 GEZM01085143 JAV60187.1 GECZ01008071 JAS61698.1 LBMM01012188 KMQ86362.1 LBMM01010405 KMQ87487.1 ABLF02041863 LBMM01012238 KMQ86334.1 ABLF02008594 GBBI01004408 JAC14304.1 GBBI01004410 JAC14302.1 LBMM01010417 KMQ87475.1 ABLF02012239 ABLF02012241 ABLF02017372 ABLF02042178 ABLF02066436 GGMR01017890 MBY30509.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 ABLF02036131 ABLF02041527 ABLF02064049 GEBQ01004229 JAT35748.1 GBHO01037676 JAG05928.1 RSAL01001808 RVE40681.1 GDHC01019528 JAP99100.1 DS497760 EFA13222.1 GAMC01021577 GAMC01021573 JAB84982.1 GEZM01054766 GEZM01054765 JAV73410.1 JAV73413.1 GEZM01060254 JAV71154.1 LBMM01009202 KMQ88311.1 ABLF02066438 QOIP01000010 RLU17691.1 ABLF02025958 GALX01000776 JAB67690.1 GBHO01034255 GBHO01034254 JAG09349.1 JAG09350.1 LBMM01009496 KMQ88090.1 GEBQ01008079 JAT31898.1 GECZ01031358 JAS38411.1 GBHO01044316 JAF99287.1 GGMR01002421 MBY15040.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 ABLF02026240 ABLF02055027 ABLF02025690 GAMC01009674 JAB96881.1 GBHO01028371 GBHO01028370 JAG15233.1 JAG15234.1 LNIX01000036 OXA39969.1 GBHO01028449 JAG15155.1 LBMM01011245 KMQ86943.1 LBMM01011244 KMQ86944.1 ABLF02067195 ABLF02039606 GBHO01008418 GBHO01008416 JAG35186.1 JAG35188.1 GGFK01004613 MBW37934.1 GGMR01005823 MBY18442.1 GBHO01009080 JAG34524.1 GGFK01004538 MBW37859.1 GGMR01000737 MBY13356.1 GBHO01029149 JAG14455.1 GGMS01014133 MBY83336.1 GGMS01009443 MBY78646.1 ABLF02025193 GGFK01004563 MBW37884.1 GGMR01003456 MBY16075.1
GBBI01004276 JAC14436.1 GBHO01044643 JAF98960.1 GEZM01054789 JAV73399.1 GAIX01013989 JAA78571.1 LBMM01008464 KMQ88855.1 GEBQ01015458 JAT24519.1 GEBQ01000825 JAT39152.1 GEBQ01028569 JAT11408.1 GDHC01006469 JAQ12160.1 GALX01004947 JAB63519.1 GEZM01085145 GEZM01085143 JAV60187.1 GECZ01008071 JAS61698.1 LBMM01012188 KMQ86362.1 LBMM01010405 KMQ87487.1 ABLF02041863 LBMM01012238 KMQ86334.1 ABLF02008594 GBBI01004408 JAC14304.1 GBBI01004410 JAC14302.1 LBMM01010417 KMQ87475.1 ABLF02012239 ABLF02012241 ABLF02017372 ABLF02042178 ABLF02066436 GGMR01017890 MBY30509.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 ABLF02036131 ABLF02041527 ABLF02064049 GEBQ01004229 JAT35748.1 GBHO01037676 JAG05928.1 RSAL01001808 RVE40681.1 GDHC01019528 JAP99100.1 DS497760 EFA13222.1 GAMC01021577 GAMC01021573 JAB84982.1 GEZM01054766 GEZM01054765 JAV73410.1 JAV73413.1 GEZM01060254 JAV71154.1 LBMM01009202 KMQ88311.1 ABLF02066438 QOIP01000010 RLU17691.1 ABLF02025958 GALX01000776 JAB67690.1 GBHO01034255 GBHO01034254 JAG09349.1 JAG09350.1 LBMM01009496 KMQ88090.1 GEBQ01008079 JAT31898.1 GECZ01031358 JAS38411.1 GBHO01044316 JAF99287.1 GGMR01002421 MBY15040.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 ABLF02026240 ABLF02055027 ABLF02025690 GAMC01009674 JAB96881.1 GBHO01028371 GBHO01028370 JAG15233.1 JAG15234.1 LNIX01000036 OXA39969.1 GBHO01028449 JAG15155.1 LBMM01011245 KMQ86943.1 LBMM01011244 KMQ86944.1 ABLF02067195 ABLF02039606 GBHO01008418 GBHO01008416 JAG35186.1 JAG35188.1 GGFK01004613 MBW37934.1 GGMR01005823 MBY18442.1 GBHO01009080 JAG34524.1 GGFK01004538 MBW37859.1 GGMR01000737 MBY13356.1 GBHO01029149 JAG14455.1 GGMS01014133 MBY83336.1 GGMS01009443 MBY78646.1 ABLF02025193 GGFK01004563 MBW37884.1 GGMR01003456 MBY16075.1
Proteomes
Pfam
Interpro
IPR008737
Peptidase_asp_put
+ More
IPR008042 Retrotrans_Pao
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR040676 DUF5641
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041588 Integrase_H2C2
IPR005312 DUF1759
IPR000477 RT_dom
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR036875 Znf_CCHC_sf
IPR001969 Aspartic_peptidase_AS
IPR017996 Royal_jelly/protein_yellow
IPR011042 6-blade_b-propeller_TolB-like
IPR029846 Yellow
IPR000727 T_SNARE_dom
IPR008042 Retrotrans_Pao
IPR036397 RNaseH_sf
IPR021109 Peptidase_aspartic_dom_sf
IPR040676 DUF5641
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041588 Integrase_H2C2
IPR005312 DUF1759
IPR000477 RT_dom
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR036875 Znf_CCHC_sf
IPR001969 Aspartic_peptidase_AS
IPR017996 Royal_jelly/protein_yellow
IPR011042 6-blade_b-propeller_TolB-like
IPR029846 Yellow
IPR000727 T_SNARE_dom
Gene 3D
ProteinModelPortal
A0A0A9Z0W7
A0A3Q0J0P0
A0A1S3DMT1
A0A0A9W266
A0A0A9VTB0
A0A023F096
+ More
A0A0A9VUS3 A0A3Q0JFQ3 A0A1Y1LIC8 A0A3Q0JJJ7 A0A1S3DM22 A0A1S3DIZ2 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A1S4EIX5 S4NIN9 A0A0J7N890 A0A1B6LLT3 A0A1B6MT91 A0A1B6KIX6 A0A1S3CUQ4 A0A3Q0IQQ3 A0A3Q0JCL2 A0A1S3DHM0 A0A146M0N4 V5GHM7 A0A1S3DH93 A0A1Y1KMX0 A0A1S3DMK8 A0A1B6GGX5 A0A0J7K7L1 A0A3Q0J7N1 A0A3Q0JBZ9 A0A3Q0J7K9 A0A0J7KB34 J9KD60 A0A0J7K7U0 A0A1S3DIE2 J9JVD6 A0A3Q0JDB5 A0A3Q0JI79 A0A1S3DHZ9 A0A023F0C4 A0A023EYR9 A0A0J7N465 A0A1S3DJJ8 X1WY64 A0A2S2PM34 J9JUM9 A0A1S3DTS8 X1WZY4 A0A1B6MIG8 A0A0A9WHD6 A0A1S3DNV9 A0A3S2TB02 A0A146KRI6 D7EKN0 W8AH72 A0A1Y1LM33 A0A1S3DBK1 A0A3Q0IZW4 A0A1Y1LIG5 A0A1Y1LBZ3 A0A0J7KDN6 X1WT50 A0A3L8DBI7 X1XU06 V5GC07 A0A0A9WWW3 A0A0J7N5Z0 A0A1B6M7K2 A0A1S3DMN9 A0A1B6EKI9 A0A0A9VYL4 A0A2S2NCY5 A0A0A9VSF7 A0A1S3DMW8 X1WVF9 X1X9U0 W8BU98 A0A0A9X8U7 A0A226D417 A0A0A9XDK6 A0A0J7K952 A0A0J7K9I5 J9K0A1 J9JUK4 A0A0A9YU87 A0A2M4AAX1 A0A2S2NNC8 A0A0A9YQQ8 A0A2M4AAS4 A0A2S2N9D0 A0A0A9XBI3 A0A2S2R014 A0A2S2QLL5 J9K351 A0A2M4AAR9 A0A2S2NFZ7
A0A0A9VUS3 A0A3Q0JFQ3 A0A1Y1LIC8 A0A3Q0JJJ7 A0A1S3DM22 A0A1S3DIZ2 A0A1S3DJ34 A0A1S3DJJ0 A0A1S3DJC0 A0A1S4EIX5 S4NIN9 A0A0J7N890 A0A1B6LLT3 A0A1B6MT91 A0A1B6KIX6 A0A1S3CUQ4 A0A3Q0IQQ3 A0A3Q0JCL2 A0A1S3DHM0 A0A146M0N4 V5GHM7 A0A1S3DH93 A0A1Y1KMX0 A0A1S3DMK8 A0A1B6GGX5 A0A0J7K7L1 A0A3Q0J7N1 A0A3Q0JBZ9 A0A3Q0J7K9 A0A0J7KB34 J9KD60 A0A0J7K7U0 A0A1S3DIE2 J9JVD6 A0A3Q0JDB5 A0A3Q0JI79 A0A1S3DHZ9 A0A023F0C4 A0A023EYR9 A0A0J7N465 A0A1S3DJJ8 X1WY64 A0A2S2PM34 J9JUM9 A0A1S3DTS8 X1WZY4 A0A1B6MIG8 A0A0A9WHD6 A0A1S3DNV9 A0A3S2TB02 A0A146KRI6 D7EKN0 W8AH72 A0A1Y1LM33 A0A1S3DBK1 A0A3Q0IZW4 A0A1Y1LIG5 A0A1Y1LBZ3 A0A0J7KDN6 X1WT50 A0A3L8DBI7 X1XU06 V5GC07 A0A0A9WWW3 A0A0J7N5Z0 A0A1B6M7K2 A0A1S3DMN9 A0A1B6EKI9 A0A0A9VYL4 A0A2S2NCY5 A0A0A9VSF7 A0A1S3DMW8 X1WVF9 X1X9U0 W8BU98 A0A0A9X8U7 A0A226D417 A0A0A9XDK6 A0A0J7K952 A0A0J7K9I5 J9K0A1 J9JUK4 A0A0A9YU87 A0A2M4AAX1 A0A2S2NNC8 A0A0A9YQQ8 A0A2M4AAS4 A0A2S2N9D0 A0A0A9XBI3 A0A2S2R014 A0A2S2QLL5 J9K351 A0A2M4AAR9 A0A2S2NFZ7
Ontologies
PANTHER
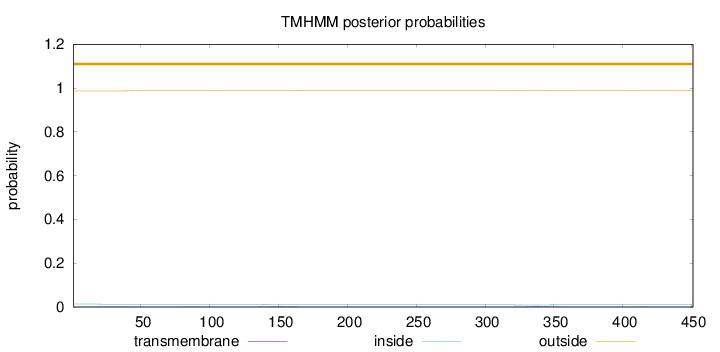
Topology
Length:
451
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12652
Exp number, first 60 AAs:
0.01465
Total prob of N-in:
0.01328
outside
1 - 451
Population Genetic Test Statistics
Pi
378.193075
Theta
190.335544
Tajima's D
3.206165
CLR
0.65813
CSRT
0.986000699965002
Interpretation
Uncertain
