Gene
KWMTBOMO10202
Pre Gene Modal
BGIBMGA005608
Annotation
PREDICTED:_glucose_dehydrogenase_[FAD?_quinone]-like_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 2.618
Sequence
CDS
ATGCTCGGTGGGAGCAGCGGTATAAATGGAATGATTTACATTAAGGGAAACTTTAAAGATTACAAAAGATGGTATGATGCAGGAAATGAGGAATGGCATCCGAACGTTGTAGAAAAATATTTTATAAAAGCAGAAAATCTTCAAGATACTGAACTATTAAAATCCGCTGAGGTTAGAAAAAATTACGGAACAAGAGGGCCTCTAATAATAAATTCATTTAACGTAACATCAAGGCGCATTGCCGATAAAATAATAGAAGCTTGGGATGAGATTGGATTCAAAAAAGTTTTGGATGTTAGTAATCTTAACAACTTACGGGGATCTGGAAGGTTTAGGGCATCTGCTGCTAGTGGTACAAGAGAAAGTACGGCTAAAGCATATTTAAATCCAATAAAAAACAGAAAAAACCTTTTCATTGTTAAGAATGCTCTTGTTACCAAGGTTTTAATAAACAATAAAACTAAGGAGGTATTTGGGGTATCTGTAGAAATTCATGGAAAAAAATTCACTTTTTATGCAAACAAGGAAGTCATTTTAAGTGCAGGTTCAATTAATACACCGCAGCTGCTGATGTTGTCTGGAGTTGGACCGCGGGAACATCTTATTTCAAAGAATATTTTTTGTAAAGTAAACTCTCGCATGGTTGGTGAAAATTTACAAGATCATGGTATAGTACTTGTTCCTGTATTTGGTAAAGAGCCGGAAGACCAAACGACAGCAGAAAAAAGTTTTGCTGCCATACAATATCTTTACAATCGTGAAGGGTACTTAGGTGGTAAAAGTACCCCGGACGTTTCAGCATTTTATTCAGAATGTGCCAACCAAACATATCCACAATTTCAAAGCGTTCCTCTTCTTTTCGGTAAAAATTCGTCATCAGTGCGTGAATTTTACAACACTATCAAAGTGAAACCTTCAGTGATAGAATCAATAGTGAAACAAAACATTAACAATGCTTTGTATATTTTTCGATTTATTTTGTTACATCCTTTTTCTAAAGGAAACATTAAGTTGCGTTCAAATGATCCAAAACAATATCCGATTATTTACGCTAATTATTTTAAAGATCCAAGGGACTTAGATGCAGCCGTAGTCGGAATTAAAATGCTGACTAAAATCGTGAATACTAAATATTTCAAATCGATTGGTGGATTTTTAGGCAGAATAGACTGGCCTCCTTGTAATAAATTCGTACTCGACACCAATGAATATTGGAGATGCATAGCTTTGAATTTTATCACCACCGTGTATCATCCAACAGGTACTACGAGAATGGGACGTAATATAAACGTGTCTGTTGTCGACAGCAGACTAAGAGTTCATGGCCTTAAAAAACTTCGCATAATAGACGCAGGTGTAATGCCATTTACAGTCAGTGCAAATACCAACGCTCCAAGTGTGATGGTGGGTGAAAGAGGAGCAGATTTGGTTAAAGAAGATTACAGAAAGTGTCCTTCAGGACGATGTCCATGTTAA
Protein
MLGGSSGINGMIYIKGNFKDYKRWYDAGNEEWHPNVVEKYFIKAENLQDTELLKSAEVRKNYGTRGPLIINSFNVTSRRIADKIIEAWDEIGFKKVLDVSNLNNLRGSGRFRASAASGTRESTAKAYLNPIKNRKNLFIVKNALVTKVLINNKTKEVFGVSVEIHGKKFTFYANKEVILSAGSINTPQLLMLSGVGPREHLISKNIFCKVNSRMVGENLQDHGIVLVPVFGKEPEDQTTAEKSFAAIQYLYNREGYLGGKSTPDVSAFYSECANQTYPQFQSVPLLFGKNSSSVREFYNTIKVKPSVIESIVKQNINNALYIFRFILLHPFSKGNIKLRSNDPKQYPIIYANYFKDPRDLDAAVVGIKMLTKIVNTKYFKSIGGFLGRIDWPPCNKFVLDTNEYWRCIALNFITTVYHPTGTTRMGRNINVSVVDSRLRVHGLKKLRIIDAGVMPFTVSANTNAPSVMVGERGADLVKEDYRKCPSGRCPC
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9J7W7
A0A0L7L9T7
A0A2A4JKJ5
A0A2H1WG27
A0A2A4JLJ2
A0A2A4IZI4
+ More
A0A2A4JW72 A0A2H1WT25 H9JYB9 A0A2H1VF08 A0A2H1VF04 A0A2H1VEJ4 A0A194QB47 A0A194QB82 A0A2H1VF60 A0A194QSF0 A0A2A4IYS9 A0A212FF54 A0A2W1B5P9 A0A2H1WZL6 A0A194QAU5 A0A0N1IJ07 A0A0L7L582 H9J861 A0A1B0GIG3 A0A1L8E0M0 A0A1L8E037 A0A0C9QE18 A0A1L8E0E7 A0A1L8DZX6 H9JTY8 A0A182SDY4 A0A182Q3I8 A0A182R5V7 A0A194PSS7 A0A2H1V0S8 A0A1Y9H2C4 A0A182MUT2 A0A2J7PXQ0 E2ABB7 F4WRC9 A0A3S2P1X7 A0A182JHR5 K7J277 A0A1B0DMM3 A0A1Q3FK91 A0A182UDY6 I4DNK7 H9JTY7 A0A1I8N6K3 A0A1L8E0T8 A0A1B6F402 B0WJG6 A0A195BBJ1 T1PFB8 A0A1B6MBY6 Q7QFX9 A0A158NUQ9 A0A151WNR9 A0A182FZT4 A0A2K8FTL4 E0VUB2 A0A0A9W6I4 A0A0N0PE61 W5J6D5 A0A1B6HJF4 E0VUA7 H9JLQ9 A0A1B6LHN9 A0A154PBS2 E0VUA8 A0A151IVV0 A0A2A4K4K5 A0A2H8TE31 A0A151J2Y2 A0A023EVK5 E2AAW1 A0A084VW89 A0A1Q3FZM7 A0A146LFQ3 A0A2J7PXM0 A0A2K8FTP1 A0A2P8ZHS7 A0A2J7RBH7 A0A151I6R9 H9JI92 A0A084VW88 A0A2P8YRH6 A0A194QNE6 A0A1Y9H2C5 F4WRC7 A0A154NWQ8 A0A0L7KNL7 A0A2W1BWE7 A0A151I6R6 A0A310SP23 A0A2J7Q2K0
A0A2A4JW72 A0A2H1WT25 H9JYB9 A0A2H1VF08 A0A2H1VF04 A0A2H1VEJ4 A0A194QB47 A0A194QB82 A0A2H1VF60 A0A194QSF0 A0A2A4IYS9 A0A212FF54 A0A2W1B5P9 A0A2H1WZL6 A0A194QAU5 A0A0N1IJ07 A0A0L7L582 H9J861 A0A1B0GIG3 A0A1L8E0M0 A0A1L8E037 A0A0C9QE18 A0A1L8E0E7 A0A1L8DZX6 H9JTY8 A0A182SDY4 A0A182Q3I8 A0A182R5V7 A0A194PSS7 A0A2H1V0S8 A0A1Y9H2C4 A0A182MUT2 A0A2J7PXQ0 E2ABB7 F4WRC9 A0A3S2P1X7 A0A182JHR5 K7J277 A0A1B0DMM3 A0A1Q3FK91 A0A182UDY6 I4DNK7 H9JTY7 A0A1I8N6K3 A0A1L8E0T8 A0A1B6F402 B0WJG6 A0A195BBJ1 T1PFB8 A0A1B6MBY6 Q7QFX9 A0A158NUQ9 A0A151WNR9 A0A182FZT4 A0A2K8FTL4 E0VUB2 A0A0A9W6I4 A0A0N0PE61 W5J6D5 A0A1B6HJF4 E0VUA7 H9JLQ9 A0A1B6LHN9 A0A154PBS2 E0VUA8 A0A151IVV0 A0A2A4K4K5 A0A2H8TE31 A0A151J2Y2 A0A023EVK5 E2AAW1 A0A084VW89 A0A1Q3FZM7 A0A146LFQ3 A0A2J7PXM0 A0A2K8FTP1 A0A2P8ZHS7 A0A2J7RBH7 A0A151I6R9 H9JI92 A0A084VW88 A0A2P8YRH6 A0A194QNE6 A0A1Y9H2C5 F4WRC7 A0A154NWQ8 A0A0L7KNL7 A0A2W1BWE7 A0A151I6R6 A0A310SP23 A0A2J7Q2K0
Pubmed
EMBL
BABH01020059
JTDY01002039
KOB72263.1
NWSH01001107
PCG72577.1
ODYU01008368
+ More
SOQ51896.1 PCG72578.1 NWSH01004383 PCG65171.1 NWSH01000521 PCG75884.1 ODYU01010827 SOQ56167.1 BABH01042896 ODYU01002217 SOQ39415.1 ODYU01002218 SOQ39418.1 ODYU01001942 SOQ38842.1 KQ459232 KPJ02687.1 KPJ02689.1 SOQ39416.1 KQ461155 KPJ08244.1 NWSH01004668 PCG64789.1 AGBW02008849 OWR52369.1 KZ150306 PZC71499.1 ODYU01012241 SOQ58442.1 KPJ02554.1 KQ460044 KPJ18321.1 JTDY01002835 KOB70627.1 BABH01039828 AJWK01014080 GFDF01002039 JAV12045.1 GFDF01002040 JAV12044.1 GBYB01012653 JAG82420.1 GFDF01002042 JAV12042.1 GFDF01002041 JAV12043.1 BABH01003799 AXCN02000040 KQ459595 KPI96023.1 ODYU01000155 SOQ34448.1 AXCM01001593 NEVH01020853 PNF21103.1 GL438237 EFN69238.1 GL888284 EGI63346.1 RSAL01000051 RVE50264.1 AAZX01008181 AJVK01016891 GFDL01007123 JAV27922.1 AK402986 BAM19497.1 BABH01003798 GFDF01001902 JAV12182.1 GECZ01024829 JAS44940.1 DS231960 EDS29162.1 KQ976532 KYM81564.1 KA646603 AFP61232.1 GEBQ01006583 JAT33394.1 AAAB01008844 EAA06000.3 ADTU01026683 KQ982905 KYQ49514.1 KY618825 AQW43011.1 DS235784 EEB16968.1 GBHO01040588 JAG03016.1 KPJ18306.1 ADMH02002107 ETN58958.1 GECU01032991 JAS74715.1 EEB16963.1 BABH01005224 BABH01005225 BABH01005226 GEBQ01016745 JAT23232.1 KQ434869 KZC09325.1 EEB16964.1 KQ980881 KYN11981.1 NWSH01000190 PCG78592.1 GFXV01000539 MBW12344.1 KQ980314 KYN16689.1 GAPW01000526 JAC13072.1 GL438191 EFN69424.1 ATLV01017521 KE525172 KFB42233.1 GFDL01002031 JAV33014.1 GDHC01012817 JAQ05812.1 PNF21089.1 KY618826 AQW43012.1 PYGN01000053 PSN56054.1 NEVH01005904 PNF38171.1 KQ978473 KYM93715.1 BABH01022249 ATLV01017518 ATLV01017519 ATLV01017520 KFB42232.1 PYGN01000411 PSN46845.1 KQ461194 KPJ06879.1 EGI63344.1 KQ434773 KZC04023.1 JTDY01008426 KOB64574.1 KZ149916 PZC77944.1 KYM93713.1 KQ759784 OAD62862.1 NEVH01019370 PNF22812.1
SOQ51896.1 PCG72578.1 NWSH01004383 PCG65171.1 NWSH01000521 PCG75884.1 ODYU01010827 SOQ56167.1 BABH01042896 ODYU01002217 SOQ39415.1 ODYU01002218 SOQ39418.1 ODYU01001942 SOQ38842.1 KQ459232 KPJ02687.1 KPJ02689.1 SOQ39416.1 KQ461155 KPJ08244.1 NWSH01004668 PCG64789.1 AGBW02008849 OWR52369.1 KZ150306 PZC71499.1 ODYU01012241 SOQ58442.1 KPJ02554.1 KQ460044 KPJ18321.1 JTDY01002835 KOB70627.1 BABH01039828 AJWK01014080 GFDF01002039 JAV12045.1 GFDF01002040 JAV12044.1 GBYB01012653 JAG82420.1 GFDF01002042 JAV12042.1 GFDF01002041 JAV12043.1 BABH01003799 AXCN02000040 KQ459595 KPI96023.1 ODYU01000155 SOQ34448.1 AXCM01001593 NEVH01020853 PNF21103.1 GL438237 EFN69238.1 GL888284 EGI63346.1 RSAL01000051 RVE50264.1 AAZX01008181 AJVK01016891 GFDL01007123 JAV27922.1 AK402986 BAM19497.1 BABH01003798 GFDF01001902 JAV12182.1 GECZ01024829 JAS44940.1 DS231960 EDS29162.1 KQ976532 KYM81564.1 KA646603 AFP61232.1 GEBQ01006583 JAT33394.1 AAAB01008844 EAA06000.3 ADTU01026683 KQ982905 KYQ49514.1 KY618825 AQW43011.1 DS235784 EEB16968.1 GBHO01040588 JAG03016.1 KPJ18306.1 ADMH02002107 ETN58958.1 GECU01032991 JAS74715.1 EEB16963.1 BABH01005224 BABH01005225 BABH01005226 GEBQ01016745 JAT23232.1 KQ434869 KZC09325.1 EEB16964.1 KQ980881 KYN11981.1 NWSH01000190 PCG78592.1 GFXV01000539 MBW12344.1 KQ980314 KYN16689.1 GAPW01000526 JAC13072.1 GL438191 EFN69424.1 ATLV01017521 KE525172 KFB42233.1 GFDL01002031 JAV33014.1 GDHC01012817 JAQ05812.1 PNF21089.1 KY618826 AQW43012.1 PYGN01000053 PSN56054.1 NEVH01005904 PNF38171.1 KQ978473 KYM93715.1 BABH01022249 ATLV01017518 ATLV01017519 ATLV01017520 KFB42232.1 PYGN01000411 PSN46845.1 KQ461194 KPJ06879.1 EGI63344.1 KQ434773 KZC04023.1 JTDY01008426 KOB64574.1 KZ149916 PZC77944.1 KYM93713.1 KQ759784 OAD62862.1 NEVH01019370 PNF22812.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000092461 UP000075901 UP000075886 UP000075884 UP000075883 UP000235965 UP000000311 UP000007755 UP000283053 UP000075880 UP000002358 UP000092462 UP000075902 UP000095301 UP000002320 UP000078540 UP000007062 UP000005205 UP000075809 UP000069272 UP000009046 UP000000673 UP000076502 UP000078492 UP000030765 UP000245037 UP000078542
UP000092461 UP000075901 UP000075886 UP000075884 UP000075883 UP000235965 UP000000311 UP000007755 UP000283053 UP000075880 UP000002358 UP000092462 UP000075902 UP000095301 UP000002320 UP000078540 UP000007062 UP000005205 UP000075809 UP000069272 UP000009046 UP000000673 UP000076502 UP000078492 UP000030765 UP000245037 UP000078542
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9J7W7
A0A0L7L9T7
A0A2A4JKJ5
A0A2H1WG27
A0A2A4JLJ2
A0A2A4IZI4
+ More
A0A2A4JW72 A0A2H1WT25 H9JYB9 A0A2H1VF08 A0A2H1VF04 A0A2H1VEJ4 A0A194QB47 A0A194QB82 A0A2H1VF60 A0A194QSF0 A0A2A4IYS9 A0A212FF54 A0A2W1B5P9 A0A2H1WZL6 A0A194QAU5 A0A0N1IJ07 A0A0L7L582 H9J861 A0A1B0GIG3 A0A1L8E0M0 A0A1L8E037 A0A0C9QE18 A0A1L8E0E7 A0A1L8DZX6 H9JTY8 A0A182SDY4 A0A182Q3I8 A0A182R5V7 A0A194PSS7 A0A2H1V0S8 A0A1Y9H2C4 A0A182MUT2 A0A2J7PXQ0 E2ABB7 F4WRC9 A0A3S2P1X7 A0A182JHR5 K7J277 A0A1B0DMM3 A0A1Q3FK91 A0A182UDY6 I4DNK7 H9JTY7 A0A1I8N6K3 A0A1L8E0T8 A0A1B6F402 B0WJG6 A0A195BBJ1 T1PFB8 A0A1B6MBY6 Q7QFX9 A0A158NUQ9 A0A151WNR9 A0A182FZT4 A0A2K8FTL4 E0VUB2 A0A0A9W6I4 A0A0N0PE61 W5J6D5 A0A1B6HJF4 E0VUA7 H9JLQ9 A0A1B6LHN9 A0A154PBS2 E0VUA8 A0A151IVV0 A0A2A4K4K5 A0A2H8TE31 A0A151J2Y2 A0A023EVK5 E2AAW1 A0A084VW89 A0A1Q3FZM7 A0A146LFQ3 A0A2J7PXM0 A0A2K8FTP1 A0A2P8ZHS7 A0A2J7RBH7 A0A151I6R9 H9JI92 A0A084VW88 A0A2P8YRH6 A0A194QNE6 A0A1Y9H2C5 F4WRC7 A0A154NWQ8 A0A0L7KNL7 A0A2W1BWE7 A0A151I6R6 A0A310SP23 A0A2J7Q2K0
A0A2A4JW72 A0A2H1WT25 H9JYB9 A0A2H1VF08 A0A2H1VF04 A0A2H1VEJ4 A0A194QB47 A0A194QB82 A0A2H1VF60 A0A194QSF0 A0A2A4IYS9 A0A212FF54 A0A2W1B5P9 A0A2H1WZL6 A0A194QAU5 A0A0N1IJ07 A0A0L7L582 H9J861 A0A1B0GIG3 A0A1L8E0M0 A0A1L8E037 A0A0C9QE18 A0A1L8E0E7 A0A1L8DZX6 H9JTY8 A0A182SDY4 A0A182Q3I8 A0A182R5V7 A0A194PSS7 A0A2H1V0S8 A0A1Y9H2C4 A0A182MUT2 A0A2J7PXQ0 E2ABB7 F4WRC9 A0A3S2P1X7 A0A182JHR5 K7J277 A0A1B0DMM3 A0A1Q3FK91 A0A182UDY6 I4DNK7 H9JTY7 A0A1I8N6K3 A0A1L8E0T8 A0A1B6F402 B0WJG6 A0A195BBJ1 T1PFB8 A0A1B6MBY6 Q7QFX9 A0A158NUQ9 A0A151WNR9 A0A182FZT4 A0A2K8FTL4 E0VUB2 A0A0A9W6I4 A0A0N0PE61 W5J6D5 A0A1B6HJF4 E0VUA7 H9JLQ9 A0A1B6LHN9 A0A154PBS2 E0VUA8 A0A151IVV0 A0A2A4K4K5 A0A2H8TE31 A0A151J2Y2 A0A023EVK5 E2AAW1 A0A084VW89 A0A1Q3FZM7 A0A146LFQ3 A0A2J7PXM0 A0A2K8FTP1 A0A2P8ZHS7 A0A2J7RBH7 A0A151I6R9 H9JI92 A0A084VW88 A0A2P8YRH6 A0A194QNE6 A0A1Y9H2C5 F4WRC7 A0A154NWQ8 A0A0L7KNL7 A0A2W1BWE7 A0A151I6R6 A0A310SP23 A0A2J7Q2K0
PDB
5OC1
E-value=2.21694e-36,
Score=383
Ontologies
PATHWAY
GO
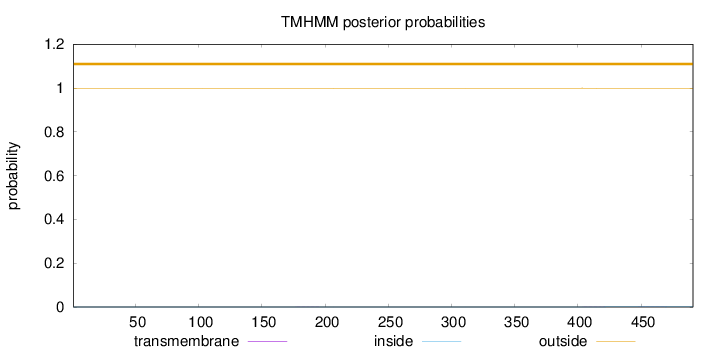
Topology
Length:
491
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01785
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00056
outside
1 - 491
Population Genetic Test Statistics
Pi
29.383991
Theta
210.641827
Tajima's D
1.907188
CLR
0
CSRT
0.865206739663017
Interpretation
Uncertain
