Gene
KWMTBOMO10201
Pre Gene Modal
BGIBMGA005609
Annotation
Uncharacterized_protein_OBRU01_12558_[Operophtera_brumata]
Location in the cell
Mitochondrial Reliability : 2.399
Sequence
CDS
ATGCTTGGTGGCAGCGGTGGTATCAACGGAATGATTTACTTTAAAGGACACGATTTTGATTTCCAACGATGGTACGACGCTGGGAACAAAGAATGGCATCCAAAAATAGTGAGATATTTTTATAAGAAACTCGAAAGTTTACAGGATGCTTCCCAGTTGCGGAATCCATTTATTAGGAGAAACTATGGCCTCAACGGCAACCTTATCATAAATAGTCTGAACAGTACAAACGAGCATTTAATAGAAAGCGTCATAAATTCCTTTGACGAAATCAGCTTCCGACGAAATTGCGATATAAGTGCTCTAAATGCGTTCGGGTCAGGAAAATTTAAAATTAACGGAAGTAATGGTAGACGAGTAAGCACAGCTAAAGCGTATTTAAACACGATACAGAAACGAAAGAATTTAAAAATTATAACAAATGCTTTTGTTACTAAAATCTTAATAGATTCTGTGACTAAAACTGTTGAAGGAGTAGAAGTAGAAGTACAAGGACAAAAGAAGACATTTTACGCAAATTTAGAAGTGATTTTGAGTGCAGGAACAATTAATACACCTCAAATATTGATGATATCAGGAATAGGTCCCGAAGATCAACTGAGTTCAAAAGGAATTTCATGCAAAGTAGATTCTCCGATGGTTGGGAAGAATCTTCAGGATCACGCTATTGTACCAGTTACAGTTTATGGTGATAAGCCAAAAAAAAAGAAGAGTGAAGCTGAACGTAATTTTGATATAATTAGATATCTATACAACCGCAGTGGCCCTTTAGCATACATTAGCTCTCCAGGCATTGCTGCTTTGTATACCGATGACCCCAATTTACCATATCCTAAATTCCAATCTTTTCTTATCTTATATGAAAAAAATACATCCTCGGTTCGCGAAGCCTTTGAAAAACGCGGTTATAAGCAATCCGTCGTAAATTCTATTTCAAAGCTAAATACTAACCACGCTCTGTATCATTTCGCGATAACACTTTTGCATCCACATTCAAAAGGTTACATTCGGTTACGTTCTAATAATCCTAAAGATCCTCCCCTAATTTACACCAACTATTTTAGCGATCCAAGAGATCTGAGATCATGCGTACGCGGTGTCAAAATGCTAACGAAAATCACAAGAACAACATATTTTAAATCTATAAATGGGTTTCTCGGTCGTATTAATTTGGAGCCGTGTAATGGTTATGAATTGGACAGTGCTGAATATTGGAAATGTATATGTTTGAATCTGGTAACGACTGTATACCATCCGGTTGGTACTTGTAAAATGGGCGTAGAACCAAAGACTTCAGTTGTGAGTAGTCGTTTGAAAGTCTTCGGTGTTGCTAAGCTTCGAATTATAGACGCTAGCGTAATGCCGAATGAAATTAGTGGAAATACAAACGCTGCTTGTGCTATGATCGGAGAAAGAGGAGCTGCAATCGTAAAGGAAGATTACTCAAAGGAAAAGCAGTACTCAATATAA
Protein
MLGGSGGINGMIYFKGHDFDFQRWYDAGNKEWHPKIVRYFYKKLESLQDASQLRNPFIRRNYGLNGNLIINSLNSTNEHLIESVINSFDEISFRRNCDISALNAFGSGKFKINGSNGRRVSTAKAYLNTIQKRKNLKIITNAFVTKILIDSVTKTVEGVEVEVQGQKKTFYANLEVILSAGTINTPQILMISGIGPEDQLSSKGISCKVDSPMVGKNLQDHAIVPVTVYGDKPKKKKSEAERNFDIIRYLYNRSGPLAYISSPGIAALYTDDPNLPYPKFQSFLILYEKNTSSVREAFEKRGYKQSVVNSISKLNTNHALYHFAITLLHPHSKGYIRLRSNNPKDPPLIYTNYFSDPRDLRSCVRGVKMLTKITRTTYFKSINGFLGRINLEPCNGYELDSAEYWKCICLNLVTTVYHPVGTCKMGVEPKTSVVSSRLKVFGVAKLRIIDASVMPNEISGNTNAACAMIGERGAAIVKEDYSKEKQYSI
Summary
Cofactor
FAD
Similarity
Belongs to the GMC oxidoreductase family.
Uniprot
H9J7W7
A0A0L7L9T7
A0A2A4JLJ2
A0A2H1WG27
A0A2A4JKJ5
A0A2A4IZI4
+ More
H9JYB9 A0A2H1VF60 A0A2H1WT25 A0A2H1VEJ4 A0A194QB47 A0A2H1VF04 A0A194QB82 A0A2H1VF08 A0A2A4JW72 A0A194QSF0 I4DJY8 A0A2A4IYS9 A0A0N1IJ07 A0A194QAU5 A0A2W1B5P9 A0A2H1WZL6 H9J861 A0A212FF54 H9JTY8 A0A182Q3I8 A0A1Y9H2C4 A0A182R5V7 A0A182MUT2 A0A1L8E0E7 A0A2H1V0S8 A0A1L8DZX6 A0A182SDY4 D2A3N1 A0A0L7LUK3 A0A182UDY6 A0A023EVK5 A0A1Q3FK91 A0A182JHR5 A0A2K8FTL4 A0A0L7KQ89 A0A182FZT4 Q7QFX9 Q17DW2 A0A0L7L582 A0A1Z2RRI5 A0A151I6I3 A0A182IM41 A0A194QNE6 W5J6D5 A0A194PPA0 A0A0C9QE18 B4JKZ8 A0A0L7KNL7 B0WJG6 E0VUB2 U5EVL2 Q17DV9 A0A023EUM5 H9JTY7 W5J850 A0A182FZT6 A0A026WCZ8 A0A182H3F4 A0A182I8U6 A0A3L8D7W0 A0A088S3T2 A0A088S3M8 A0A1B6D0X9 A0A182LAR7 A0A195BAI7 F5HJA6 A0A1Q3FZM7 A0A1B0DMM3 A0A182JP92 A0A1B0GIG3 A0A151K0E9 B0WJG7 E9IKG5 B0W3I1 A0A088S376 A0A088SM07 A0A2M4AHF2 A0A336LLP9 A0A2P8ZHS7 A0A158NUR3 A0A1W4X2S9 A0A151J323 E2ABB7 A0A1Y9H2C6 N6T1Q2 E2AAW0 A0A154PBR8 A0A2M4AFT0 A0A0J7KLX4 A0A2M4AF63 T1PFB8 A0A084VW88 A0A088S5A0
H9JYB9 A0A2H1VF60 A0A2H1WT25 A0A2H1VEJ4 A0A194QB47 A0A2H1VF04 A0A194QB82 A0A2H1VF08 A0A2A4JW72 A0A194QSF0 I4DJY8 A0A2A4IYS9 A0A0N1IJ07 A0A194QAU5 A0A2W1B5P9 A0A2H1WZL6 H9J861 A0A212FF54 H9JTY8 A0A182Q3I8 A0A1Y9H2C4 A0A182R5V7 A0A182MUT2 A0A1L8E0E7 A0A2H1V0S8 A0A1L8DZX6 A0A182SDY4 D2A3N1 A0A0L7LUK3 A0A182UDY6 A0A023EVK5 A0A1Q3FK91 A0A182JHR5 A0A2K8FTL4 A0A0L7KQ89 A0A182FZT4 Q7QFX9 Q17DW2 A0A0L7L582 A0A1Z2RRI5 A0A151I6I3 A0A182IM41 A0A194QNE6 W5J6D5 A0A194PPA0 A0A0C9QE18 B4JKZ8 A0A0L7KNL7 B0WJG6 E0VUB2 U5EVL2 Q17DV9 A0A023EUM5 H9JTY7 W5J850 A0A182FZT6 A0A026WCZ8 A0A182H3F4 A0A182I8U6 A0A3L8D7W0 A0A088S3T2 A0A088S3M8 A0A1B6D0X9 A0A182LAR7 A0A195BAI7 F5HJA6 A0A1Q3FZM7 A0A1B0DMM3 A0A182JP92 A0A1B0GIG3 A0A151K0E9 B0WJG7 E9IKG5 B0W3I1 A0A088S376 A0A088SM07 A0A2M4AHF2 A0A336LLP9 A0A2P8ZHS7 A0A158NUR3 A0A1W4X2S9 A0A151J323 E2ABB7 A0A1Y9H2C6 N6T1Q2 E2AAW0 A0A154PBR8 A0A2M4AFT0 A0A0J7KLX4 A0A2M4AF63 T1PFB8 A0A084VW88 A0A088S5A0
Pubmed
EMBL
BABH01020059
JTDY01002039
KOB72263.1
NWSH01001107
PCG72578.1
ODYU01008368
+ More
SOQ51896.1 PCG72577.1 NWSH01004383 PCG65171.1 BABH01042896 ODYU01002217 SOQ39416.1 ODYU01010827 SOQ56167.1 ODYU01001942 SOQ38842.1 KQ459232 KPJ02687.1 ODYU01002218 SOQ39418.1 KPJ02689.1 SOQ39415.1 NWSH01000521 PCG75884.1 KQ461155 KPJ08244.1 AK401606 BAM18228.1 NWSH01004668 PCG64789.1 KQ460044 KPJ18321.1 KPJ02554.1 KZ150306 PZC71499.1 ODYU01012241 SOQ58442.1 BABH01039828 AGBW02008849 OWR52369.1 BABH01003799 AXCN02000040 AXCM01001593 GFDF01002042 JAV12042.1 ODYU01000155 SOQ34448.1 GFDF01002041 JAV12043.1 KQ971348 EFA05531.1 JTDY01000066 KOB79079.1 GAPW01000526 JAC13072.1 GFDL01007123 JAV27922.1 KY618825 AQW43011.1 JTDY01007102 KOB65457.1 AAAB01008844 EAA06000.3 CH477290 EAT44639.1 JTDY01002835 KOB70627.1 KY938814 ASA46455.1 KQ978473 KYM93710.1 KQ461194 KPJ06879.1 ADMH02002107 ETN58958.1 KQ459603 KPI92960.1 GBYB01012653 JAG82420.1 CH916370 EDW00251.1 JTDY01008426 KOB64574.1 DS231960 EDS29162.1 DS235784 EEB16968.1 GANO01001829 JAB58042.1 EAT44642.1 GAPW01000947 JAC12651.1 BABH01003798 ETN58959.1 KK107261 EZA53937.1 JXUM01024562 KQ560694 KXJ81238.1 APCN01003121 QOIP01000012 RLU16172.1 KF717661 AIO02863.1 KF717664 AIO02866.1 GEDC01017950 JAS19348.1 KQ976532 KYM81561.1 EGK96367.1 EGK96368.1 GFDL01002031 JAV33014.1 AJVK01016891 AJWK01014080 KQ981296 KYN43080.1 EDS29163.1 GL763984 EFZ18935.1 DS231832 EDS31767.1 KF717658 AIO02860.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 GGFK01006898 MBW40219.1 UFQS01000011 UFQT01000011 SSW97203.1 SSX17589.1 PYGN01000053 PSN56054.1 ADTU01026685 KQ980314 KYN16692.1 GL438237 EFN69238.1 APGK01055703 APGK01055704 KB741269 ENN71448.1 GL438191 EFN69423.1 KQ434869 KZC09326.1 GGFK01006251 MBW39572.1 LBMM01005695 KMQ91287.1 GGFK01006105 MBW39426.1 KA646603 AFP61232.1 ATLV01017518 ATLV01017519 ATLV01017520 ATLV01017521 KE525172 KFB42232.1 KF717655 AIO02857.1
SOQ51896.1 PCG72577.1 NWSH01004383 PCG65171.1 BABH01042896 ODYU01002217 SOQ39416.1 ODYU01010827 SOQ56167.1 ODYU01001942 SOQ38842.1 KQ459232 KPJ02687.1 ODYU01002218 SOQ39418.1 KPJ02689.1 SOQ39415.1 NWSH01000521 PCG75884.1 KQ461155 KPJ08244.1 AK401606 BAM18228.1 NWSH01004668 PCG64789.1 KQ460044 KPJ18321.1 KPJ02554.1 KZ150306 PZC71499.1 ODYU01012241 SOQ58442.1 BABH01039828 AGBW02008849 OWR52369.1 BABH01003799 AXCN02000040 AXCM01001593 GFDF01002042 JAV12042.1 ODYU01000155 SOQ34448.1 GFDF01002041 JAV12043.1 KQ971348 EFA05531.1 JTDY01000066 KOB79079.1 GAPW01000526 JAC13072.1 GFDL01007123 JAV27922.1 KY618825 AQW43011.1 JTDY01007102 KOB65457.1 AAAB01008844 EAA06000.3 CH477290 EAT44639.1 JTDY01002835 KOB70627.1 KY938814 ASA46455.1 KQ978473 KYM93710.1 KQ461194 KPJ06879.1 ADMH02002107 ETN58958.1 KQ459603 KPI92960.1 GBYB01012653 JAG82420.1 CH916370 EDW00251.1 JTDY01008426 KOB64574.1 DS231960 EDS29162.1 DS235784 EEB16968.1 GANO01001829 JAB58042.1 EAT44642.1 GAPW01000947 JAC12651.1 BABH01003798 ETN58959.1 KK107261 EZA53937.1 JXUM01024562 KQ560694 KXJ81238.1 APCN01003121 QOIP01000012 RLU16172.1 KF717661 AIO02863.1 KF717664 AIO02866.1 GEDC01017950 JAS19348.1 KQ976532 KYM81561.1 EGK96367.1 EGK96368.1 GFDL01002031 JAV33014.1 AJVK01016891 AJWK01014080 KQ981296 KYN43080.1 EDS29163.1 GL763984 EFZ18935.1 DS231832 EDS31767.1 KF717658 AIO02860.1 KF717656 KF717657 KF717660 KF717667 AIO02858.1 AIO02859.1 AIO02862.1 AIO02869.1 GGFK01006898 MBW40219.1 UFQS01000011 UFQT01000011 SSW97203.1 SSX17589.1 PYGN01000053 PSN56054.1 ADTU01026685 KQ980314 KYN16692.1 GL438237 EFN69238.1 APGK01055703 APGK01055704 KB741269 ENN71448.1 GL438191 EFN69423.1 KQ434869 KZC09326.1 GGFK01006251 MBW39572.1 LBMM01005695 KMQ91287.1 GGFK01006105 MBW39426.1 KA646603 AFP61232.1 ATLV01017518 ATLV01017519 ATLV01017520 ATLV01017521 KE525172 KFB42232.1 KF717655 AIO02857.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000075886 UP000075884 UP000075883 UP000075901 UP000007266 UP000075902 UP000075880 UP000069272 UP000007062 UP000008820 UP000078542 UP000000673 UP000001070 UP000002320 UP000009046 UP000053097 UP000069940 UP000249989 UP000075840 UP000279307 UP000075882 UP000078540 UP000092462 UP000075881 UP000092461 UP000078541 UP000245037 UP000005205 UP000192223 UP000078492 UP000000311 UP000019118 UP000076502 UP000036403 UP000030765
UP000075886 UP000075884 UP000075883 UP000075901 UP000007266 UP000075902 UP000075880 UP000069272 UP000007062 UP000008820 UP000078542 UP000000673 UP000001070 UP000002320 UP000009046 UP000053097 UP000069940 UP000249989 UP000075840 UP000279307 UP000075882 UP000078540 UP000092462 UP000075881 UP000092461 UP000078541 UP000245037 UP000005205 UP000192223 UP000078492 UP000000311 UP000019118 UP000076502 UP000036403 UP000030765
Interpro
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9J7W7
A0A0L7L9T7
A0A2A4JLJ2
A0A2H1WG27
A0A2A4JKJ5
A0A2A4IZI4
+ More
H9JYB9 A0A2H1VF60 A0A2H1WT25 A0A2H1VEJ4 A0A194QB47 A0A2H1VF04 A0A194QB82 A0A2H1VF08 A0A2A4JW72 A0A194QSF0 I4DJY8 A0A2A4IYS9 A0A0N1IJ07 A0A194QAU5 A0A2W1B5P9 A0A2H1WZL6 H9J861 A0A212FF54 H9JTY8 A0A182Q3I8 A0A1Y9H2C4 A0A182R5V7 A0A182MUT2 A0A1L8E0E7 A0A2H1V0S8 A0A1L8DZX6 A0A182SDY4 D2A3N1 A0A0L7LUK3 A0A182UDY6 A0A023EVK5 A0A1Q3FK91 A0A182JHR5 A0A2K8FTL4 A0A0L7KQ89 A0A182FZT4 Q7QFX9 Q17DW2 A0A0L7L582 A0A1Z2RRI5 A0A151I6I3 A0A182IM41 A0A194QNE6 W5J6D5 A0A194PPA0 A0A0C9QE18 B4JKZ8 A0A0L7KNL7 B0WJG6 E0VUB2 U5EVL2 Q17DV9 A0A023EUM5 H9JTY7 W5J850 A0A182FZT6 A0A026WCZ8 A0A182H3F4 A0A182I8U6 A0A3L8D7W0 A0A088S3T2 A0A088S3M8 A0A1B6D0X9 A0A182LAR7 A0A195BAI7 F5HJA6 A0A1Q3FZM7 A0A1B0DMM3 A0A182JP92 A0A1B0GIG3 A0A151K0E9 B0WJG7 E9IKG5 B0W3I1 A0A088S376 A0A088SM07 A0A2M4AHF2 A0A336LLP9 A0A2P8ZHS7 A0A158NUR3 A0A1W4X2S9 A0A151J323 E2ABB7 A0A1Y9H2C6 N6T1Q2 E2AAW0 A0A154PBR8 A0A2M4AFT0 A0A0J7KLX4 A0A2M4AF63 T1PFB8 A0A084VW88 A0A088S5A0
H9JYB9 A0A2H1VF60 A0A2H1WT25 A0A2H1VEJ4 A0A194QB47 A0A2H1VF04 A0A194QB82 A0A2H1VF08 A0A2A4JW72 A0A194QSF0 I4DJY8 A0A2A4IYS9 A0A0N1IJ07 A0A194QAU5 A0A2W1B5P9 A0A2H1WZL6 H9J861 A0A212FF54 H9JTY8 A0A182Q3I8 A0A1Y9H2C4 A0A182R5V7 A0A182MUT2 A0A1L8E0E7 A0A2H1V0S8 A0A1L8DZX6 A0A182SDY4 D2A3N1 A0A0L7LUK3 A0A182UDY6 A0A023EVK5 A0A1Q3FK91 A0A182JHR5 A0A2K8FTL4 A0A0L7KQ89 A0A182FZT4 Q7QFX9 Q17DW2 A0A0L7L582 A0A1Z2RRI5 A0A151I6I3 A0A182IM41 A0A194QNE6 W5J6D5 A0A194PPA0 A0A0C9QE18 B4JKZ8 A0A0L7KNL7 B0WJG6 E0VUB2 U5EVL2 Q17DV9 A0A023EUM5 H9JTY7 W5J850 A0A182FZT6 A0A026WCZ8 A0A182H3F4 A0A182I8U6 A0A3L8D7W0 A0A088S3T2 A0A088S3M8 A0A1B6D0X9 A0A182LAR7 A0A195BAI7 F5HJA6 A0A1Q3FZM7 A0A1B0DMM3 A0A182JP92 A0A1B0GIG3 A0A151K0E9 B0WJG7 E9IKG5 B0W3I1 A0A088S376 A0A088SM07 A0A2M4AHF2 A0A336LLP9 A0A2P8ZHS7 A0A158NUR3 A0A1W4X2S9 A0A151J323 E2ABB7 A0A1Y9H2C6 N6T1Q2 E2AAW0 A0A154PBR8 A0A2M4AFT0 A0A0J7KLX4 A0A2M4AF63 T1PFB8 A0A084VW88 A0A088S5A0
PDB
5OC1
E-value=3.82487e-36,
Score=381
Ontologies
GO
Topology
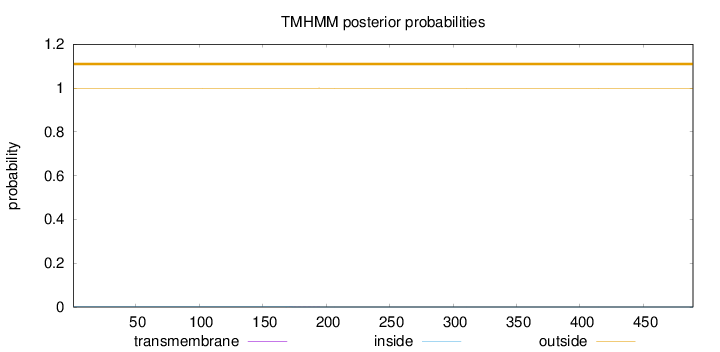
Length:
489
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01083
Exp number, first 60 AAs:
0.00016
Total prob of N-in:
0.00082
outside
1 - 489
Population Genetic Test Statistics
Pi
196.400557
Theta
199.192218
Tajima's D
0.116247
CLR
0.001252
CSRT
0.396030198490075
Interpretation
Uncertain
