Gene
KWMTBOMO10200
Pre Gene Modal
BGIBMGA005611
Annotation
PREDICTED:_protein_eva-1-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.829 Nuclear Reliability : 1.949
Sequence
CDS
ATGGTGACACTCGTGTGTCCCCGTGGAACAACCATTAGCATCCAGGTAGCGCAGTACGGAGCTTCAGTAACCCAAGGATCCTGTGCCTCAGAGCTGGCCGAGTATCAACCGGTGGCAGTTGAAGTTATTGGCGACACATCCTGCGTCTGGCCTAGTACTCTACAGTATTCCTTGTTGCAGACAGTTGTAGAGGCATGTCAGAAGAAGCGACAGTGCAAGTTCCACACCAGTCCCAAGGCGTTCGGAATGGACCCATGCCCCGGTTCCAGAAGGTTCGTCGAGGTCGCCTATAAGTGCCGTCCTTATGAATTTCGAAGTAAAGTGGGATGTGAGAACGACGTGTTACACTTGAGCTGCAACCCCCACTCGAGGGTCGCTATTTATTCCGCTCAGTATGGCAGGACGGAGTATGACTCGATACAGTGCCCTCAACCGCGCGGCATGAAAGAAGAAACCTGCCTGGAGCCCTATGCCACAGAAAAAGCGATGCGAGAATGCCACGGCAAGAGGAGGTGCGTTCTATCCGCCGACAGCAATATGTTCGGGAATCCTTGCCGACCCGGCAGCAGAACTTATCTCAAAGTGGTTTACACTTGCGTTCCGCGTACAGTTCTCAAAGAGAGGTACGAAAGTGCCCCTGAAGAGGACGAGGCTGCTCACGATGTTTCCGATCCCGAGCACGAAGATGTCGACGAATCAAACGACCGATGGTGGGGCGAATCAGCAGTTCCTCCGGCTCCAGCAGTAGCAGCTGTACCCCCTCAACGCCCTTCTACGACGACCGTCTTAAACGCAACCAGCATAAATAGGATCGAACCTCCAGCCGTACCAACGAATCATCACTTATCTAAAGATGATAGCTTCGACGTAATATACGTCTATATAATAGCGGCTGTGGCTGTGGCATTGCTACTCTGCATCATCGTTGGGATCATCCGCTGCGTTATGCATAAACAAAGCACTGACCAGCCCAAAGGGCCCGACGTGTCTGCCACAACTGAAATCCCCAACGGATTCAACGACAGCATCTCCGAGGTAGACAACGACATCAACATAACAAGTCTTTCCGGTCCGGTCGATTCCGCTGATTCCGGAATGAAACAAGATATGCAATTTGGGAATGTGGCTGTCAGTCCCAAAATCAATAGGTACGTCGGTAGGCCAGTTCCTAACACATATCCTCACGTCAGTACTAATATGTATGGGCAGGTTGCAGAATTCCCAATAGAAATGCCACTGCGGACTATACCGATGCACGGTACTTTAGGCCGAGGAGTTGCGGTCAAAACACTACCGAGAGTCCAAGTGCAATCAGAAATAGACCCTAACACGAGAACTGAAAAGGATACCATTAAAAAGCCAGCCGAGAGAAACGAAAAACTCGAACGCTACAAACGTAAAACAATTAAGAGGAACTAA
Protein
MVTLVCPRGTTISIQVAQYGASVTQGSCASELAEYQPVAVEVIGDTSCVWPSTLQYSLLQTVVEACQKKRQCKFHTSPKAFGMDPCPGSRRFVEVAYKCRPYEFRSKVGCENDVLHLSCNPHSRVAIYSAQYGRTEYDSIQCPQPRGMKEETCLEPYATEKAMRECHGKRRCVLSADSNMFGNPCRPGSRTYLKVVYTCVPRTVLKERYESAPEEDEAAHDVSDPEHEDVDESNDRWWGESAVPPAPAVAAVPPQRPSTTTVLNATSINRIEPPAVPTNHHLSKDDSFDVIYVYIIAAVAVALLLCIIVGIIRCVMHKQSTDQPKGPDVSATTEIPNGFNDSISEVDNDINITSLSGPVDSADSGMKQDMQFGNVAVSPKINRYVGRPVPNTYPHVSTNMYGQVAEFPIEMPLRTIPMHGTLGRGVAVKTLPRVQVQSEIDPNTRTEKDTIKKPAERNEKLERYKRKTIKRN
Summary
Uniprot
A0A2A4JVS6
A0A194QCI0
A0A194QXE0
A0A1Y1NAX1
A0A1Y1NC36
A0A194QSE5
+ More
A0A194QB52 Q17F45 B0WUZ4 A0A195F230 A0A2H1VDG0 A0A0Q9WH53 A0A2H8TLG1 B4JRG1 B4LXG1 A0A2A3EDE0 A0A2A4JVK7 B4NIV7 A0A1I8MWL7 A0A3S2LYM4 B4GM87 B5DVP2 A0A0R3NJ94 A0A1W4VS28 A0A0P8ZDR8 A0A1W4VDR9 A0A212EN37 A0A2A4JXC7 A0A1B0BXE3 A0A034V5A3 A0A0K8VKG5 A0A0K8VRK9 A0A0K8W6B2 A0A0R1DZS1
A0A194QB52 Q17F45 B0WUZ4 A0A195F230 A0A2H1VDG0 A0A0Q9WH53 A0A2H8TLG1 B4JRG1 B4LXG1 A0A2A3EDE0 A0A2A4JVK7 B4NIV7 A0A1I8MWL7 A0A3S2LYM4 B4GM87 B5DVP2 A0A0R3NJ94 A0A1W4VS28 A0A0P8ZDR8 A0A1W4VDR9 A0A212EN37 A0A2A4JXC7 A0A1B0BXE3 A0A034V5A3 A0A0K8VKG5 A0A0K8VRK9 A0A0K8W6B2 A0A0R1DZS1
Pubmed
EMBL
NWSH01000521
PCG75886.1
KQ459232
KPJ02690.1
KQ461155
KPJ08241.1
+ More
GEZM01010838 JAV93785.1 GEZM01010837 JAV93786.1 KPJ08239.1 KPJ02692.1 CH477276 EAT45138.1 DS232113 EDS35316.1 KQ981856 KYN34441.1 ODYU01001960 SOQ38878.1 CH940650 KRF83506.1 GFXV01002313 MBW14118.1 CH916373 EDV94351.1 EDW67839.2 KRF83507.1 KZ288275 PBC29728.1 NWSH01000490 PCG76041.1 CH964272 EDW84859.2 RSAL01000110 RVE47158.1 CH479185 EDW37961.1 CM000070 EDY68450.3 KRT01047.1 KRT01048.1 CH902623 KPU72703.1 AGBW02013741 OWR42896.1 PCG76042.1 JXJN01022193 GAKP01020476 JAC38476.1 GDHF01012947 JAI39367.1 GDHF01010836 JAI41478.1 GDHF01026094 GDHF01021343 GDHF01008615 GDHF01005720 GDHF01000761 JAI26220.1 JAI30971.1 JAI43699.1 JAI46594.1 JAI51553.1 CM000160 KRK02557.1
GEZM01010838 JAV93785.1 GEZM01010837 JAV93786.1 KPJ08239.1 KPJ02692.1 CH477276 EAT45138.1 DS232113 EDS35316.1 KQ981856 KYN34441.1 ODYU01001960 SOQ38878.1 CH940650 KRF83506.1 GFXV01002313 MBW14118.1 CH916373 EDV94351.1 EDW67839.2 KRF83507.1 KZ288275 PBC29728.1 NWSH01000490 PCG76041.1 CH964272 EDW84859.2 RSAL01000110 RVE47158.1 CH479185 EDW37961.1 CM000070 EDY68450.3 KRT01047.1 KRT01048.1 CH902623 KPU72703.1 AGBW02013741 OWR42896.1 PCG76042.1 JXJN01022193 GAKP01020476 JAC38476.1 GDHF01012947 JAI39367.1 GDHF01010836 JAI41478.1 GDHF01026094 GDHF01021343 GDHF01008615 GDHF01005720 GDHF01000761 JAI26220.1 JAI30971.1 JAI43699.1 JAI46594.1 JAI51553.1 CM000160 KRK02557.1
Proteomes
Pfam
PF02140 Gal_Lectin
Interpro
IPR000922
Lectin_gal-bd_dom
ProteinModelPortal
A0A2A4JVS6
A0A194QCI0
A0A194QXE0
A0A1Y1NAX1
A0A1Y1NC36
A0A194QSE5
+ More
A0A194QB52 Q17F45 B0WUZ4 A0A195F230 A0A2H1VDG0 A0A0Q9WH53 A0A2H8TLG1 B4JRG1 B4LXG1 A0A2A3EDE0 A0A2A4JVK7 B4NIV7 A0A1I8MWL7 A0A3S2LYM4 B4GM87 B5DVP2 A0A0R3NJ94 A0A1W4VS28 A0A0P8ZDR8 A0A1W4VDR9 A0A212EN37 A0A2A4JXC7 A0A1B0BXE3 A0A034V5A3 A0A0K8VKG5 A0A0K8VRK9 A0A0K8W6B2 A0A0R1DZS1
A0A194QB52 Q17F45 B0WUZ4 A0A195F230 A0A2H1VDG0 A0A0Q9WH53 A0A2H8TLG1 B4JRG1 B4LXG1 A0A2A3EDE0 A0A2A4JVK7 B4NIV7 A0A1I8MWL7 A0A3S2LYM4 B4GM87 B5DVP2 A0A0R3NJ94 A0A1W4VS28 A0A0P8ZDR8 A0A1W4VDR9 A0A212EN37 A0A2A4JXC7 A0A1B0BXE3 A0A034V5A3 A0A0K8VKG5 A0A0K8VRK9 A0A0K8W6B2 A0A0R1DZS1
PDB
2ZX4
E-value=1.17195e-14,
Score=195
Ontologies
GO
Topology
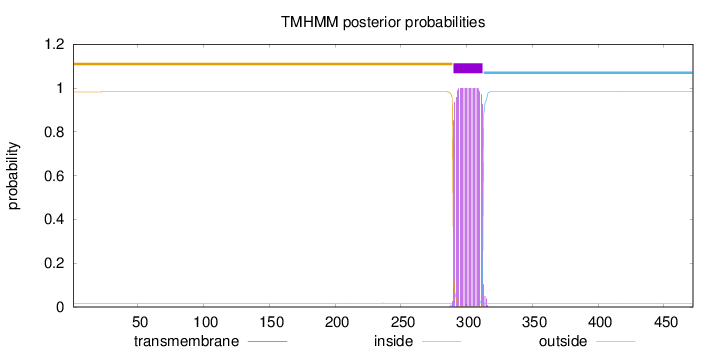
Length:
472
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.85711
Exp number, first 60 AAs:
0.000780000000000001
Total prob of N-in:
0.01751
outside
1 - 289
TMhelix
290 - 312
inside
313 - 472
Population Genetic Test Statistics
Pi
204.519147
Theta
161.752527
Tajima's D
0.807821
CLR
0.394849
CSRT
0.605069746512674
Interpretation
Uncertain
