Gene
KWMTBOMO10198
Pre Gene Modal
BGIBMGA000463
Annotation
aminopeptidase_N_precursor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.876
Sequence
CDS
ATGAAGAAGCTATGGTCTGCAGAAGGTTACACTTGGGGATACCAAATTTACAATGGTCAAAGTGTGCAAGTATTTGGTTTGGATACCGGAAGTATAGTAGTAACATTGGCAGAAGGTTTACTTGATGAAGGTCGCTGCATGATAACTGACAACTACTACACGAGCGACCCTTTGACAGAATTTATGTTTTCTCGCAATACAGACCTCTGTGGCACAGTCAATAAAAAGAGAAGAGGTTTACCGAAAGAGAGAAGGAGAGAAAAGCTGAGCATTCTAGCAGCACATGAAACCATTGTAAAAAAACTTTTATAG
Protein
MKKLWSAEGYTWGYQIYNGQSVQVFGLDTGSIVVTLAEGLLDEGRCMITDNYYTSDPLTEFMFSRNTDLCGTVNKKRRGLPKERRREKLSILAAHETIVKKLL
Summary
Similarity
Belongs to the universal ribosomal protein uS3 family.
Uniprot
H9IT83
A0A1E1VXQ8
A0A212FPU3
J9JPM8
A0A0D6LDM3
A0A016SIZ8
+ More
H9J315 A0A0C2DK98 A0A1S3DK31 A0A0P4WIX9 A0A0N4YIJ5 A0A0N4XY30 A0A0N4UUN0 A0A016TZU4 A0A016U0C6 A0A016V127 A0A0N4YQB3 A0A016U6U0 A0A0P4W9M6 A0A016WV22 A0A016VJ95 S4PBU0 A0A0D6LMR8 A0A087SXJ9 A0A1A9VDM4 A0A131YKY1 A0A0N4XI68 A0A1E1WBV4 J9LK68 A0A016VFC3 A0A1B6HG03 A0A016VFZ3 A0A2H1W1W2 B7PQY6 A0A016T3Q5 A0A0D6LHL4 A0A1B6HS81 A0A016VEZ7 A0A2J7PPC7 A0A2J7PER0 A0A0D6M5K7 E2AF83 T0MM39 A0A2J7PCA5 A0A1B6JZ80 A0A147BEZ3 A0A182YRA2 A0A1B6IUZ3 A0A0N7ZCZ9 J9K106 X1WS81 L7M1S0 C7ENH0 A0A183G7U2 A0A3P8A9A8 S4P945 A0A2J7QSN3 A0A183GK12 A0A3P8GNV7 A0A1B6ISI9 A0A0L8HTK5 A0A3B4B068 A0A2S2QUS6 A0A2H1VNZ4 A0A0N4XUZ6 A0A2S2R8C3 A0A1B6M084 A0A3B4AZ14 X1WK26 J9JVX5 W4ZHM1 A0A2J7PL02 A0A1B6JCM5 A0A3Q3A6G3 A0A2J7PPH7 S4PBM2 A0A2J7R0C5 A0A368GTV8 A0A2J7PNH7 S4NY54 J9KGK0 A0A023F0B8 A0A023EYQ2 A0A2S2R5V8 A0A131Y2F1 A0A147BL95 J9LWR0 A0A3Q0JNJ9 S4P9J6 A0A0N4WYZ7 E2C9Y4 A8J4K9 W6NB59
H9J315 A0A0C2DK98 A0A1S3DK31 A0A0P4WIX9 A0A0N4YIJ5 A0A0N4XY30 A0A0N4UUN0 A0A016TZU4 A0A016U0C6 A0A016V127 A0A0N4YQB3 A0A016U6U0 A0A0P4W9M6 A0A016WV22 A0A016VJ95 S4PBU0 A0A0D6LMR8 A0A087SXJ9 A0A1A9VDM4 A0A131YKY1 A0A0N4XI68 A0A1E1WBV4 J9LK68 A0A016VFC3 A0A1B6HG03 A0A016VFZ3 A0A2H1W1W2 B7PQY6 A0A016T3Q5 A0A0D6LHL4 A0A1B6HS81 A0A016VEZ7 A0A2J7PPC7 A0A2J7PER0 A0A0D6M5K7 E2AF83 T0MM39 A0A2J7PCA5 A0A1B6JZ80 A0A147BEZ3 A0A182YRA2 A0A1B6IUZ3 A0A0N7ZCZ9 J9K106 X1WS81 L7M1S0 C7ENH0 A0A183G7U2 A0A3P8A9A8 S4P945 A0A2J7QSN3 A0A183GK12 A0A3P8GNV7 A0A1B6ISI9 A0A0L8HTK5 A0A3B4B068 A0A2S2QUS6 A0A2H1VNZ4 A0A0N4XUZ6 A0A2S2R8C3 A0A1B6M084 A0A3B4AZ14 X1WK26 J9JVX5 W4ZHM1 A0A2J7PL02 A0A1B6JCM5 A0A3Q3A6G3 A0A2J7PPH7 S4PBM2 A0A2J7R0C5 A0A368GTV8 A0A2J7PNH7 S4NY54 J9KGK0 A0A023F0B8 A0A023EYQ2 A0A2S2R5V8 A0A131Y2F1 A0A147BL95 J9LWR0 A0A3Q0JNJ9 S4P9J6 A0A0N4WYZ7 E2C9Y4 A8J4K9 W6NB59
Pubmed
EMBL
BABH01013489
GDQN01011536
GDQN01001322
JAT79518.1
JAT89732.1
AGBW02000393
+ More
OWR55781.1 ABLF02017387 KE125214 EPB70205.1 JARK01001552 EYB90668.1 BABH01041459 KN728983 KIH62942.1 GDRN01069744 JAI63996.1 UYSL01022372 VDL80334.1 UYSL01019948 VDL71548.1 UXUI01007140 VDD85673.1 JARK01001402 EYC08311.1 EYC08312.1 JARK01001356 EYC20951.1 UYSL01024178 VDL83170.1 JARK01001390 EYC10641.1 GDRN01075668 JAI63019.1 JARK01000104 EYC43082.1 JARK01001344 EYC27674.1 GAIX01005747 JAA86813.1 KE125003 EPB73164.1 KK112412 KFM57588.1 GEDV01009986 JAP78571.1 UYSL01002382 VDL65810.1 GDQN01006584 JAT84470.1 ABLF02011361 JARK01001346 EYC26339.1 GECU01034165 JAS73541.1 EYC26340.1 ODYU01005788 SOQ47017.1 ABJB010539332 DS768824 EEC09008.1 JARK01001477 EYB97365.1 KE125300 EPB69401.1 GECU01030201 JAS77505.1 EYC26204.1 NEVH01023279 PNF18149.1 NEVH01026090 PNF14815.1 KE124806 EPB78763.1 GL439060 EFN67906.1 KE647018 EQB62075.1 NEVH01027063 NEVH01021202 NEVH01002692 PNF13962.1 PNF19936.1 PNF41621.1 GECU01003202 JAT04505.1 GEGO01006047 JAR89357.1 GECU01016947 JAS90759.1 GDRN01056216 JAI65856.1 ABLF02038619 ABLF02010857 ABLF02042012 GACK01007981 JAA57053.1 GQ281011 ACT79642.1 UZAH01030316 VDP10112.1 GAIX01005691 JAA86869.1 NEVH01011876 PNF31583.1 UZAH01034601 VDP36123.1 GECU01017853 JAS89853.1 KQ417398 KOF92090.1 GGMS01012057 MBY81260.1 ODYU01003590 SOQ42555.1 UYSL01019810 VDL70162.1 GGMS01016409 MBY85612.1 GEBQ01010639 JAT29338.1 ABLF02014546 ABLF02014549 ABLF02042029 ABLF02030622 ABLF02030627 AAGJ04151167 NEVH01024529 PNF17014.1 GECU01010818 JAS96888.1 NEVH01022661 PNF18242.1 GAIX01004551 JAA88009.1 NEVH01008275 PNF34265.1 JOJR01000056 RCN47811.1 NEVH01023953 PNF17885.1 GAIX01013935 JAA78625.1 ABLF02008544 GBBI01004438 JAC14274.1 GBBI01004440 JAC14272.1 GGMS01016193 MBY85396.1 GEFM01002122 JAP73674.1 GEGO01004329 JAR91075.1 ABLF02029982 GAIX01003729 JAA88831.1 UZAF01019782 VDO63509.1 GL453910 EFN75248.1 AB300558 BAF82021.1 CAVP010055412 CDL94120.1
OWR55781.1 ABLF02017387 KE125214 EPB70205.1 JARK01001552 EYB90668.1 BABH01041459 KN728983 KIH62942.1 GDRN01069744 JAI63996.1 UYSL01022372 VDL80334.1 UYSL01019948 VDL71548.1 UXUI01007140 VDD85673.1 JARK01001402 EYC08311.1 EYC08312.1 JARK01001356 EYC20951.1 UYSL01024178 VDL83170.1 JARK01001390 EYC10641.1 GDRN01075668 JAI63019.1 JARK01000104 EYC43082.1 JARK01001344 EYC27674.1 GAIX01005747 JAA86813.1 KE125003 EPB73164.1 KK112412 KFM57588.1 GEDV01009986 JAP78571.1 UYSL01002382 VDL65810.1 GDQN01006584 JAT84470.1 ABLF02011361 JARK01001346 EYC26339.1 GECU01034165 JAS73541.1 EYC26340.1 ODYU01005788 SOQ47017.1 ABJB010539332 DS768824 EEC09008.1 JARK01001477 EYB97365.1 KE125300 EPB69401.1 GECU01030201 JAS77505.1 EYC26204.1 NEVH01023279 PNF18149.1 NEVH01026090 PNF14815.1 KE124806 EPB78763.1 GL439060 EFN67906.1 KE647018 EQB62075.1 NEVH01027063 NEVH01021202 NEVH01002692 PNF13962.1 PNF19936.1 PNF41621.1 GECU01003202 JAT04505.1 GEGO01006047 JAR89357.1 GECU01016947 JAS90759.1 GDRN01056216 JAI65856.1 ABLF02038619 ABLF02010857 ABLF02042012 GACK01007981 JAA57053.1 GQ281011 ACT79642.1 UZAH01030316 VDP10112.1 GAIX01005691 JAA86869.1 NEVH01011876 PNF31583.1 UZAH01034601 VDP36123.1 GECU01017853 JAS89853.1 KQ417398 KOF92090.1 GGMS01012057 MBY81260.1 ODYU01003590 SOQ42555.1 UYSL01019810 VDL70162.1 GGMS01016409 MBY85612.1 GEBQ01010639 JAT29338.1 ABLF02014546 ABLF02014549 ABLF02042029 ABLF02030622 ABLF02030627 AAGJ04151167 NEVH01024529 PNF17014.1 GECU01010818 JAS96888.1 NEVH01022661 PNF18242.1 GAIX01004551 JAA88009.1 NEVH01008275 PNF34265.1 JOJR01000056 RCN47811.1 NEVH01023953 PNF17885.1 GAIX01013935 JAA78625.1 ABLF02008544 GBBI01004438 JAC14274.1 GBBI01004440 JAC14272.1 GGMS01016193 MBY85396.1 GEFM01002122 JAP73674.1 GEGO01004329 JAR91075.1 ABLF02029982 GAIX01003729 JAA88831.1 UZAF01019782 VDO63509.1 GL453910 EFN75248.1 AB300558 BAF82021.1 CAVP010055412 CDL94120.1
Proteomes
UP000005204
UP000007151
UP000007819
UP000024635
UP000079169
UP000038043
+ More
UP000271162 UP000038041 UP000274131 UP000054359 UP000078200 UP000001555 UP000235965 UP000000311 UP000076408 UP000050761 UP000053454 UP000261520 UP000007110 UP000264800 UP000252519 UP000038042 UP000268014 UP000008237 UP000025227
UP000271162 UP000038041 UP000274131 UP000054359 UP000078200 UP000001555 UP000235965 UP000000311 UP000076408 UP000050761 UP000053454 UP000261520 UP000007110 UP000264800 UP000252519 UP000038042 UP000268014 UP000008237 UP000025227
Pfam
Interpro
IPR029526
PGBD
+ More
IPR008042 Retrotrans_Pao
IPR003034 SAP_dom
IPR036361 SAP_dom_sf
IPR032718 PGBD4_C_Znf-ribbon
IPR014044 CAP_domain
IPR035940 CAP_sf
IPR000337 GPCR_3
IPR028082 Peripla_BP_I
IPR001828 ANF_lig-bd_rcpt
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR008906 HATC_C_dom
IPR012337 RNaseH-like_sf
IPR005703 Ribosomal_S3_euk/arc
IPR036419 Ribosomal_S3_C_sf
IPR001351 Ribosomal_S3_C
IPR004044 KH_dom_type_2
IPR018280 Ribosomal_S3_CS
IPR015946 KH_dom-like_a/b
IPR009019 KH_sf_prok-type
IPR008042 Retrotrans_Pao
IPR003034 SAP_dom
IPR036361 SAP_dom_sf
IPR032718 PGBD4_C_Znf-ribbon
IPR014044 CAP_domain
IPR035940 CAP_sf
IPR000337 GPCR_3
IPR028082 Peripla_BP_I
IPR001828 ANF_lig-bd_rcpt
IPR036691 Endo/exonu/phosph_ase_sf
IPR000477 RT_dom
IPR008906 HATC_C_dom
IPR012337 RNaseH-like_sf
IPR005703 Ribosomal_S3_euk/arc
IPR036419 Ribosomal_S3_C_sf
IPR001351 Ribosomal_S3_C
IPR004044 KH_dom_type_2
IPR018280 Ribosomal_S3_CS
IPR015946 KH_dom-like_a/b
IPR009019 KH_sf_prok-type
SUPFAM
ProteinModelPortal
H9IT83
A0A1E1VXQ8
A0A212FPU3
J9JPM8
A0A0D6LDM3
A0A016SIZ8
+ More
H9J315 A0A0C2DK98 A0A1S3DK31 A0A0P4WIX9 A0A0N4YIJ5 A0A0N4XY30 A0A0N4UUN0 A0A016TZU4 A0A016U0C6 A0A016V127 A0A0N4YQB3 A0A016U6U0 A0A0P4W9M6 A0A016WV22 A0A016VJ95 S4PBU0 A0A0D6LMR8 A0A087SXJ9 A0A1A9VDM4 A0A131YKY1 A0A0N4XI68 A0A1E1WBV4 J9LK68 A0A016VFC3 A0A1B6HG03 A0A016VFZ3 A0A2H1W1W2 B7PQY6 A0A016T3Q5 A0A0D6LHL4 A0A1B6HS81 A0A016VEZ7 A0A2J7PPC7 A0A2J7PER0 A0A0D6M5K7 E2AF83 T0MM39 A0A2J7PCA5 A0A1B6JZ80 A0A147BEZ3 A0A182YRA2 A0A1B6IUZ3 A0A0N7ZCZ9 J9K106 X1WS81 L7M1S0 C7ENH0 A0A183G7U2 A0A3P8A9A8 S4P945 A0A2J7QSN3 A0A183GK12 A0A3P8GNV7 A0A1B6ISI9 A0A0L8HTK5 A0A3B4B068 A0A2S2QUS6 A0A2H1VNZ4 A0A0N4XUZ6 A0A2S2R8C3 A0A1B6M084 A0A3B4AZ14 X1WK26 J9JVX5 W4ZHM1 A0A2J7PL02 A0A1B6JCM5 A0A3Q3A6G3 A0A2J7PPH7 S4PBM2 A0A2J7R0C5 A0A368GTV8 A0A2J7PNH7 S4NY54 J9KGK0 A0A023F0B8 A0A023EYQ2 A0A2S2R5V8 A0A131Y2F1 A0A147BL95 J9LWR0 A0A3Q0JNJ9 S4P9J6 A0A0N4WYZ7 E2C9Y4 A8J4K9 W6NB59
H9J315 A0A0C2DK98 A0A1S3DK31 A0A0P4WIX9 A0A0N4YIJ5 A0A0N4XY30 A0A0N4UUN0 A0A016TZU4 A0A016U0C6 A0A016V127 A0A0N4YQB3 A0A016U6U0 A0A0P4W9M6 A0A016WV22 A0A016VJ95 S4PBU0 A0A0D6LMR8 A0A087SXJ9 A0A1A9VDM4 A0A131YKY1 A0A0N4XI68 A0A1E1WBV4 J9LK68 A0A016VFC3 A0A1B6HG03 A0A016VFZ3 A0A2H1W1W2 B7PQY6 A0A016T3Q5 A0A0D6LHL4 A0A1B6HS81 A0A016VEZ7 A0A2J7PPC7 A0A2J7PER0 A0A0D6M5K7 E2AF83 T0MM39 A0A2J7PCA5 A0A1B6JZ80 A0A147BEZ3 A0A182YRA2 A0A1B6IUZ3 A0A0N7ZCZ9 J9K106 X1WS81 L7M1S0 C7ENH0 A0A183G7U2 A0A3P8A9A8 S4P945 A0A2J7QSN3 A0A183GK12 A0A3P8GNV7 A0A1B6ISI9 A0A0L8HTK5 A0A3B4B068 A0A2S2QUS6 A0A2H1VNZ4 A0A0N4XUZ6 A0A2S2R8C3 A0A1B6M084 A0A3B4AZ14 X1WK26 J9JVX5 W4ZHM1 A0A2J7PL02 A0A1B6JCM5 A0A3Q3A6G3 A0A2J7PPH7 S4PBM2 A0A2J7R0C5 A0A368GTV8 A0A2J7PNH7 S4NY54 J9KGK0 A0A023F0B8 A0A023EYQ2 A0A2S2R5V8 A0A131Y2F1 A0A147BL95 J9LWR0 A0A3Q0JNJ9 S4P9J6 A0A0N4WYZ7 E2C9Y4 A8J4K9 W6NB59
Ontologies
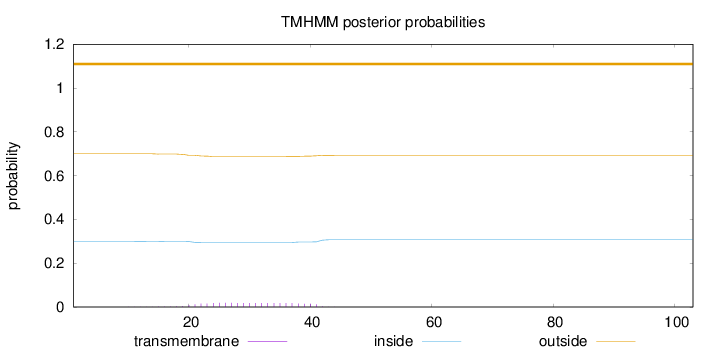
Topology
Length:
103
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.382
Exp number, first 60 AAs:
0.382
Total prob of N-in:
0.30022
outside
1 - 103
Population Genetic Test Statistics
Pi
298.432858
Theta
213.665462
Tajima's D
1.263266
CLR
0
CSRT
0.730663466826659
Interpretation
Uncertain
