Gene
KWMTBOMO10193
Pre Gene Modal
BGIBMGA005613
Annotation
PREDICTED:_transmembrane_protein_104_homolog_[Bombyx_mori]
Full name
Transmembrane protein 104 homolog
Location in the cell
PlasmaMembrane Reliability : 4.964
Sequence
CDS
ATGCCTATAACTGAGACTGGGGATCAATATTCGATATGGGTGGGCTTAATATATATATTCAATTTGATTGTTGGGACTGGCGCTCTGACATTACCTGCAGCATTTTCCAGGGCAGGATGGGCATTAAGCACAATATCATTAGTCTTCTTAGCATTTATGAGTTTCTTGAATGCAACTTATGTGATAGAAACTATGGCATGCGCCAATGCAGTGCTAAAATGGAAGAAACTGCAGTCTATTAAGAGAGAGAGTATCCAAGATAGGGATACTGATGAAGAGACTACCTCACAACAATATGGAGATCTCGAAGAACCCCTTGTTTGTGGTACTTCCATACCTTCAAGATACTACAGTCTGGACCAACGTGTGGAACTTGGAGAAATGGCAAACCTTTTTTTTAGCAAGACTGGAAGGTTGATGTTTTATTGCACTCTCTGTGTATATCTGTATGGAGACCTATCGATATATTCAGCTGCAGTTGCTAAGTCCGTTATGGATGTTGTATGTTCAACAGTCTCTTCGAATGTCACGAACGGCAGAGGTTGGGAGAGCTTGCCGTGTTTCAACGCAACCGATGATCCGATGGCGTACACTCGACTCGATGTGTATAGGGTCTCTCTGCTCACCTTCGTAGCGGTCATGGGGCCCTTCGTCTTCTTCAACGTGCAGAAGACTAAGTACCTGCAGTTGCTGACGTCAGGAATGAGATGGCTCGCATTCACGATAATGATCTCAATGGCGATCCATCTGTTAACGTCCCGAGGGGCTGAAGGCAGACCTCCTACGTTTGACGTGACGGGAATGCCCACATTATTTGGAGCATGTGTGTATTCATTCATGTGCCACCACTCGTTGCCAGGACTCATCAGTCCGATACGAGGCAAGAACGGCTTGGGGCTCCATCTCTCTCTAGATTACGTTCTCATAAGCGTATTCTACCTGTTATTAGCGTTCACTGGCGCTTTCGCCTTTGCCGAACTAAACGATTTGTACACGTTAAATTTTGTGCCGTCGGACAATGAAAACATATTCCTTAAAATTGTCGATTATTTCCTTTCACTGTTCCCGGTTTTCACTCTGTCCACGAGTTTCCCGATTATAGCGATTACATTAAAGAACAATCTGCAGTCTCTATGTTTGGACACGGGTCGCATTGAAAGTTACAATGTTGTCGTTAGAAAATTTTTATTTCCCGTCGTTACGGTAGTTCCTCCGGTACTGTTGACGTATTATTTAGAGGATATCAGTGTGTTGATTGAGTTCACTGGTTCGTATGCGGGAGCCGGTATTCAATATTTGATACCGACGTTTCTCGTTGTGTCCGCCAGGCGGCACTGCGTCGCCCTGCTGGGTTTGGGCGTGGTCAATAAGTATAAGAGTCCGTTCTCGCATGCGGCGTGGGCAGTATTCGTCCTGCTCTGGTCGTGCATGTGTATAATATTGGTCTCCGTGCACATATTAGAAAAGAAATCATGA
Protein
MPITETGDQYSIWVGLIYIFNLIVGTGALTLPAAFSRAGWALSTISLVFLAFMSFLNATYVIETMACANAVLKWKKLQSIKRESIQDRDTDEETTSQQYGDLEEPLVCGTSIPSRYYSLDQRVELGEMANLFFSKTGRLMFYCTLCVYLYGDLSIYSAAVAKSVMDVVCSTVSSNVTNGRGWESLPCFNATDDPMAYTRLDVYRVSLLTFVAVMGPFVFFNVQKTKYLQLLTSGMRWLAFTIMISMAIHLLTSRGAEGRPPTFDVTGMPTLFGACVYSFMCHHSLPGLISPIRGKNGLGLHLSLDYVLISVFYLLLAFTGAFAFAELNDLYTLNFVPSDNENIFLKIVDYFLSLFPVFTLSTSFPIIAITLKNNLQSLCLDTGRIESYNVVVRKFLFPVVTVVPPVLLTYYLEDISVLIEFTGSYAGAGIQYLIPTFLVVSARRHCVALLGLGVVNKYKSPFSHAAWAVFVLLWSCMCIILVSVHILEKKS
Summary
Similarity
Belongs to the TMEM104 family.
Keywords
Complete proteome
Glycoprotein
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Transmembrane protein 104 homolog
Uniprot
H9J7X1
A0A2H1WLN8
A0A2A4JI46
A0A194QCH0
A0A212F1T6
A0A2Z5U7P0
+ More
A0A194QXF0 A0A023ETA1 A0A182HDD4 A0A0L7QNB3 M9PD31 Q9VPF8 A0A182VRF4 A0A182K4B4 A0A182XW16 A0A151JNK6 Q0IGE5 A0A158NV73 E2BJ04 A0A182S9U9 U5EYG0 B4QRZ2 B4PF84 A0A151K1G9 B4IUG2 A0A182PE15 B0XHE2 F4WEV8 A0A182LUY9 A0A1W4W6T7 A0A182HZW7 B3NIL8 E2AFQ6 B4IAA6 A0A182XGF9 A0A182VBV0 Q7PYT6 A0A182TSD9 K7J5G5 A0A1Q3F561 A0A026WMZ1 A0A182QB65 A0A195CUP9 A0A232FLP5 A0A182N6B4 A0A151X9U8 A0A182RT66 A0A087ZZF4 A0A182IMK4 B3M995 W5J5W2 A0A2M4APJ0 A0A182F7Z8 A0A2A3EM52 A0A2M4BK33 B0XHE4 A0A2M4BK58 A0A3B0JZ43 Q2LYK0 B4HBU6 D6WGR5 A0A1Y1N8E0 T1P912 A0A1I8MY39 A0A1I8MY26 A0A1I8NQ41 A0A1I8NQ37 A0A0L0CDM1 A0A067RWK9 B4MXR5 B4MF29 A0A310SAC3 D0Z7D1 A0A0T6BCW2 A0A2J7PLX5 J3JZF2 N6TNU0 A0A1S3HNE6 A0A131Y596 A0A1W4WYM3 A0A1L8DEM4 A0A3Q2XM93 A0A3B5QMX8 A0A1S3HPU1 A0A3Q3K974 A0A3Q4GUL4 A0A2U9CTF6 A0A3Q1J7M4 A0A3P8QIT1 A0A3P9ATD4 A0A3P9L8U3 A0A3Q2XA18 A0A3B4FSQ3 A0A3Q3MP50 A0A3B3C558 A0A3P9HL30 A0A3B4VMT7 I3JSZ8 H2L5J0 A0A3B3VP50
A0A194QXF0 A0A023ETA1 A0A182HDD4 A0A0L7QNB3 M9PD31 Q9VPF8 A0A182VRF4 A0A182K4B4 A0A182XW16 A0A151JNK6 Q0IGE5 A0A158NV73 E2BJ04 A0A182S9U9 U5EYG0 B4QRZ2 B4PF84 A0A151K1G9 B4IUG2 A0A182PE15 B0XHE2 F4WEV8 A0A182LUY9 A0A1W4W6T7 A0A182HZW7 B3NIL8 E2AFQ6 B4IAA6 A0A182XGF9 A0A182VBV0 Q7PYT6 A0A182TSD9 K7J5G5 A0A1Q3F561 A0A026WMZ1 A0A182QB65 A0A195CUP9 A0A232FLP5 A0A182N6B4 A0A151X9U8 A0A182RT66 A0A087ZZF4 A0A182IMK4 B3M995 W5J5W2 A0A2M4APJ0 A0A182F7Z8 A0A2A3EM52 A0A2M4BK33 B0XHE4 A0A2M4BK58 A0A3B0JZ43 Q2LYK0 B4HBU6 D6WGR5 A0A1Y1N8E0 T1P912 A0A1I8MY39 A0A1I8MY26 A0A1I8NQ41 A0A1I8NQ37 A0A0L0CDM1 A0A067RWK9 B4MXR5 B4MF29 A0A310SAC3 D0Z7D1 A0A0T6BCW2 A0A2J7PLX5 J3JZF2 N6TNU0 A0A1S3HNE6 A0A131Y596 A0A1W4WYM3 A0A1L8DEM4 A0A3Q2XM93 A0A3B5QMX8 A0A1S3HPU1 A0A3Q3K974 A0A3Q4GUL4 A0A2U9CTF6 A0A3Q1J7M4 A0A3P8QIT1 A0A3P9ATD4 A0A3P9L8U3 A0A3Q2XA18 A0A3B4FSQ3 A0A3Q3MP50 A0A3B3C558 A0A3P9HL30 A0A3B4VMT7 I3JSZ8 H2L5J0 A0A3B3VP50
Pubmed
19121390
26354079
22118469
24945155
26483478
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 25244985 17510324 21347285 20798317 17994087 22936249 17550304 21719571 12364791 14747013 17210077 20075255 24508170 30249741 28648823 20920257 23761445 15632085 18362917 19820115 28004739 25315136 26108605 24845553 22516182 23537049 23542700 25186727 17554307 29451363
12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 25244985 17510324 21347285 20798317 17994087 22936249 17550304 21719571 12364791 14747013 17210077 20075255 24508170 30249741 28648823 20920257 23761445 15632085 18362917 19820115 28004739 25315136 26108605 24845553 22516182 23537049 23542700 25186727 17554307 29451363
EMBL
BABH01020114
BABH01020115
ODYU01009425
SOQ53842.1
NWSH01001429
PCG71254.1
+ More
KQ459232 KPJ02680.1 AGBW02010829 OWR47673.1 AP017548 BBB06958.1 KQ461155 KPJ08251.1 GAPW01001075 JAC12523.1 JXUM01001649 JXUM01001650 KQ414855 KOC60132.1 AE014296 AGB94802.1 AY118576 KQ978820 KYN28145.1 CH477192 EAT48593.1 ADTU01026935 GL448543 EFN84349.1 GANO01000380 JAB59491.1 CM000363 CM002912 EDX11238.1 KMZ00785.1 CM000159 EDW95170.1 KQ981189 KYN45236.1 CH891902 EDX00026.1 DS233146 EDS28355.1 GL888111 EGI67275.1 AXCM01003940 APCN01002017 CH954178 EDV52514.1 GL439124 EFN67735.1 CH480826 EDW44219.1 AAAB01008987 EAA01001.5 GFDL01012437 JAV22608.1 KK107167 QOIP01000010 EZA56474.1 RLU17999.1 AXCN02000281 KQ977279 KYN04385.1 NNAY01000065 OXU31378.1 KQ982365 KYQ57142.1 CH902618 EDV40079.2 ADMH02002036 ETN59832.1 GGFK01009368 MBW42689.1 KZ288212 PBC32798.1 GGFJ01004295 MBW53436.1 EDS28357.1 GGFJ01004296 MBW53437.1 OUUW01000012 SPP87347.1 CH379069 EAL29862.1 CH479267 EDW39540.1 KQ971327 EEZ99619.1 GEZM01010252 JAV94123.1 KA645187 AFP59816.1 JRES01000536 KNC30312.1 KK852421 KDR24294.1 CH963876 EDW76834.1 CH940665 EDW71130.2 KQ779356 OAD51991.1 CM000831 ACY70515.1 LJIG01001748 KRT85192.1 NEVH01024423 PNF17331.1 BT128634 AEE63591.1 APGK01057450 APGK01057451 KB741280 KB632033 ENN70910.1 ERL88214.1 GEFM01002134 JAP73662.1 GFDF01009196 JAV04888.1 CP026261 AWP19403.1 AERX01008320 AERX01008321 AERX01008322 AERX01008323
KQ459232 KPJ02680.1 AGBW02010829 OWR47673.1 AP017548 BBB06958.1 KQ461155 KPJ08251.1 GAPW01001075 JAC12523.1 JXUM01001649 JXUM01001650 KQ414855 KOC60132.1 AE014296 AGB94802.1 AY118576 KQ978820 KYN28145.1 CH477192 EAT48593.1 ADTU01026935 GL448543 EFN84349.1 GANO01000380 JAB59491.1 CM000363 CM002912 EDX11238.1 KMZ00785.1 CM000159 EDW95170.1 KQ981189 KYN45236.1 CH891902 EDX00026.1 DS233146 EDS28355.1 GL888111 EGI67275.1 AXCM01003940 APCN01002017 CH954178 EDV52514.1 GL439124 EFN67735.1 CH480826 EDW44219.1 AAAB01008987 EAA01001.5 GFDL01012437 JAV22608.1 KK107167 QOIP01000010 EZA56474.1 RLU17999.1 AXCN02000281 KQ977279 KYN04385.1 NNAY01000065 OXU31378.1 KQ982365 KYQ57142.1 CH902618 EDV40079.2 ADMH02002036 ETN59832.1 GGFK01009368 MBW42689.1 KZ288212 PBC32798.1 GGFJ01004295 MBW53436.1 EDS28357.1 GGFJ01004296 MBW53437.1 OUUW01000012 SPP87347.1 CH379069 EAL29862.1 CH479267 EDW39540.1 KQ971327 EEZ99619.1 GEZM01010252 JAV94123.1 KA645187 AFP59816.1 JRES01000536 KNC30312.1 KK852421 KDR24294.1 CH963876 EDW76834.1 CH940665 EDW71130.2 KQ779356 OAD51991.1 CM000831 ACY70515.1 LJIG01001748 KRT85192.1 NEVH01024423 PNF17331.1 BT128634 AEE63591.1 APGK01057450 APGK01057451 KB741280 KB632033 ENN70910.1 ERL88214.1 GEFM01002134 JAP73662.1 GFDF01009196 JAV04888.1 CP026261 AWP19403.1 AERX01008320 AERX01008321 AERX01008322 AERX01008323
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000069940
+ More
UP000053825 UP000000803 UP000075920 UP000075881 UP000076408 UP000078492 UP000008820 UP000005205 UP000008237 UP000075901 UP000000304 UP000002282 UP000078541 UP000075885 UP000002320 UP000007755 UP000075883 UP000192221 UP000075840 UP000008711 UP000000311 UP000001292 UP000076407 UP000075903 UP000007062 UP000075902 UP000002358 UP000053097 UP000279307 UP000075886 UP000078542 UP000215335 UP000075884 UP000075809 UP000075900 UP000005203 UP000075880 UP000007801 UP000000673 UP000069272 UP000242457 UP000268350 UP000001819 UP000008744 UP000007266 UP000095301 UP000095300 UP000037069 UP000027135 UP000007798 UP000008792 UP000235965 UP000019118 UP000030742 UP000085678 UP000192223 UP000264820 UP000002852 UP000261600 UP000261580 UP000246464 UP000265040 UP000265100 UP000265160 UP000265180 UP000264840 UP000261460 UP000261640 UP000261560 UP000265200 UP000261420 UP000005207 UP000001038 UP000261500
UP000053825 UP000000803 UP000075920 UP000075881 UP000076408 UP000078492 UP000008820 UP000005205 UP000008237 UP000075901 UP000000304 UP000002282 UP000078541 UP000075885 UP000002320 UP000007755 UP000075883 UP000192221 UP000075840 UP000008711 UP000000311 UP000001292 UP000076407 UP000075903 UP000007062 UP000075902 UP000002358 UP000053097 UP000279307 UP000075886 UP000078542 UP000215335 UP000075884 UP000075809 UP000075900 UP000005203 UP000075880 UP000007801 UP000000673 UP000069272 UP000242457 UP000268350 UP000001819 UP000008744 UP000007266 UP000095301 UP000095300 UP000037069 UP000027135 UP000007798 UP000008792 UP000235965 UP000019118 UP000030742 UP000085678 UP000192223 UP000264820 UP000002852 UP000261600 UP000261580 UP000246464 UP000265040 UP000265100 UP000265160 UP000265180 UP000264840 UP000261460 UP000261640 UP000261560 UP000265200 UP000261420 UP000005207 UP000001038 UP000261500
Pfam
PF01490 Aa_trans
Interpro
IPR013057
AA_transpt_TM
ProteinModelPortal
H9J7X1
A0A2H1WLN8
A0A2A4JI46
A0A194QCH0
A0A212F1T6
A0A2Z5U7P0
+ More
A0A194QXF0 A0A023ETA1 A0A182HDD4 A0A0L7QNB3 M9PD31 Q9VPF8 A0A182VRF4 A0A182K4B4 A0A182XW16 A0A151JNK6 Q0IGE5 A0A158NV73 E2BJ04 A0A182S9U9 U5EYG0 B4QRZ2 B4PF84 A0A151K1G9 B4IUG2 A0A182PE15 B0XHE2 F4WEV8 A0A182LUY9 A0A1W4W6T7 A0A182HZW7 B3NIL8 E2AFQ6 B4IAA6 A0A182XGF9 A0A182VBV0 Q7PYT6 A0A182TSD9 K7J5G5 A0A1Q3F561 A0A026WMZ1 A0A182QB65 A0A195CUP9 A0A232FLP5 A0A182N6B4 A0A151X9U8 A0A182RT66 A0A087ZZF4 A0A182IMK4 B3M995 W5J5W2 A0A2M4APJ0 A0A182F7Z8 A0A2A3EM52 A0A2M4BK33 B0XHE4 A0A2M4BK58 A0A3B0JZ43 Q2LYK0 B4HBU6 D6WGR5 A0A1Y1N8E0 T1P912 A0A1I8MY39 A0A1I8MY26 A0A1I8NQ41 A0A1I8NQ37 A0A0L0CDM1 A0A067RWK9 B4MXR5 B4MF29 A0A310SAC3 D0Z7D1 A0A0T6BCW2 A0A2J7PLX5 J3JZF2 N6TNU0 A0A1S3HNE6 A0A131Y596 A0A1W4WYM3 A0A1L8DEM4 A0A3Q2XM93 A0A3B5QMX8 A0A1S3HPU1 A0A3Q3K974 A0A3Q4GUL4 A0A2U9CTF6 A0A3Q1J7M4 A0A3P8QIT1 A0A3P9ATD4 A0A3P9L8U3 A0A3Q2XA18 A0A3B4FSQ3 A0A3Q3MP50 A0A3B3C558 A0A3P9HL30 A0A3B4VMT7 I3JSZ8 H2L5J0 A0A3B3VP50
A0A194QXF0 A0A023ETA1 A0A182HDD4 A0A0L7QNB3 M9PD31 Q9VPF8 A0A182VRF4 A0A182K4B4 A0A182XW16 A0A151JNK6 Q0IGE5 A0A158NV73 E2BJ04 A0A182S9U9 U5EYG0 B4QRZ2 B4PF84 A0A151K1G9 B4IUG2 A0A182PE15 B0XHE2 F4WEV8 A0A182LUY9 A0A1W4W6T7 A0A182HZW7 B3NIL8 E2AFQ6 B4IAA6 A0A182XGF9 A0A182VBV0 Q7PYT6 A0A182TSD9 K7J5G5 A0A1Q3F561 A0A026WMZ1 A0A182QB65 A0A195CUP9 A0A232FLP5 A0A182N6B4 A0A151X9U8 A0A182RT66 A0A087ZZF4 A0A182IMK4 B3M995 W5J5W2 A0A2M4APJ0 A0A182F7Z8 A0A2A3EM52 A0A2M4BK33 B0XHE4 A0A2M4BK58 A0A3B0JZ43 Q2LYK0 B4HBU6 D6WGR5 A0A1Y1N8E0 T1P912 A0A1I8MY39 A0A1I8MY26 A0A1I8NQ41 A0A1I8NQ37 A0A0L0CDM1 A0A067RWK9 B4MXR5 B4MF29 A0A310SAC3 D0Z7D1 A0A0T6BCW2 A0A2J7PLX5 J3JZF2 N6TNU0 A0A1S3HNE6 A0A131Y596 A0A1W4WYM3 A0A1L8DEM4 A0A3Q2XM93 A0A3B5QMX8 A0A1S3HPU1 A0A3Q3K974 A0A3Q4GUL4 A0A2U9CTF6 A0A3Q1J7M4 A0A3P8QIT1 A0A3P9ATD4 A0A3P9L8U3 A0A3Q2XA18 A0A3B4FSQ3 A0A3Q3MP50 A0A3B3C558 A0A3P9HL30 A0A3B4VMT7 I3JSZ8 H2L5J0 A0A3B3VP50
Ontologies
GO
Topology
Subcellular location
Membrane
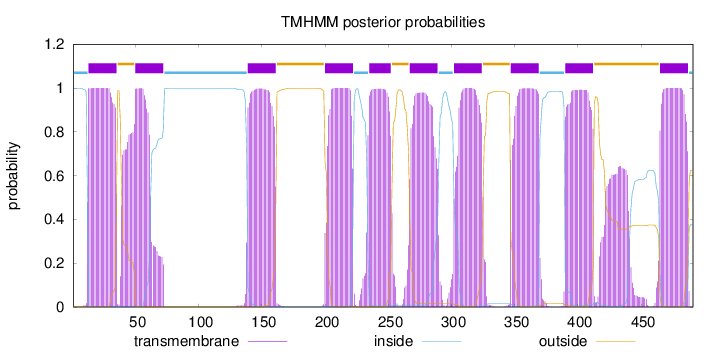
Length:
491
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
233.71686
Exp number, first 60 AAs:
41.65455
Total prob of N-in:
0.99898
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 49
TMhelix
50 - 72
inside
73 - 138
TMhelix
139 - 161
outside
162 - 199
TMhelix
200 - 222
inside
223 - 234
TMhelix
235 - 252
outside
253 - 266
TMhelix
267 - 289
inside
290 - 301
TMhelix
302 - 324
outside
325 - 346
TMhelix
347 - 369
inside
370 - 389
TMhelix
390 - 412
outside
413 - 464
TMhelix
465 - 487
inside
488 - 491
Population Genetic Test Statistics
Pi
241.297962
Theta
144.312789
Tajima's D
1.000301
CLR
0.163202
CSRT
0.658767061646918
Interpretation
Uncertain
