Pre Gene Modal
BGIBMGA005588
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_E2/E3_hybrid_ubiquitin-protein_ligase_UBE2O_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.271 Mitochondrial Reliability : 1.171
Sequence
CDS
ATGGAAGCGGAAGTGGAGGGCGCCCCTGTCTCCGGGGGCGAGGGGTTTGCGCTGCTGGAGCTCGCGCCGGCGGCGCACCGCTTCCGCCTCTCCATACTGCAGCCGTCGGACCCGCGCTCCTTCCACTCGGCCGTCAAACGAGAGATCAAATTGCTCAGGAGTGATCTACCACCAGGCGTCTGGGTCCGCGGCTACGAGAACCGCATCGATCTGCTGTCGGTGATGATAGCGGGCCCCGCGAACACGCCGTACGAGGACGGGCTGTTCGTGTTCGACGTGCAGCTCGGCGGGGAGTACCCGCGCGCGCCGCCGCTCTGCCACTACCACTCGTACTGCACGGACCGACTCAACCCGAACCTGTACGAGGACGGGAAGGTTTGTGTGTCTCTCCTGGGCACGTGGTCTGGGCGCGGTGTCGAGGTGTGGGGCAAAGACAGCTCACTCCTACAGGTCATAGTGTCTCTGCAGGGACTGATCCTCAATGCCGAGCCTTATTTCAACGAGGCTGGATATGAGAAACAAAAAGGTACCCAACAAGGCAAGGAGAACTCGCGCATGTACAACGAGATGGTGCTGTTAAAACTCGTTCAATCGATGACGAAGATGGTCCTTAACCCGCCGGAACCGTTCCGCGACGAGATCATGCAGCACATGCGCACGCACGCCGCCGCGCTCTGTGCGCGCCTCGACGGACTCGTGGCGCTCTCCGGGGAGGCGCCCCTCCCCCGCCCCGTCACGCCCCCGGACTACCCCCTGGTGCCGGCGTCGCGCGGCTTCTGCCTCACGCTCAGGTCGTCCCTCGAAGCGTTCCGAGCGGCGCTGCGTAGGAACGACATCACTCTCCCCCCGGAGGATTTATAG
Protein
MEAEVEGAPVSGGEGFALLELAPAAHRFRLSILQPSDPRSFHSAVKREIKLLRSDLPPGVWVRGYENRIDLLSVMIAGPANTPYEDGLFVFDVQLGGEYPRAPPLCHYHSYCTDRLNPNLYEDGKVCVSLLGTWSGRGVEVWGKDSSLLQVIVSLQGLILNAEPYFNEAGYEKQKGTQQGKENSRMYNEMVLLKLVQSMTKMVLNPPEPFRDEIMQHMRTHAAALCARLDGLVALSGEAPLPRPVTPPDYPLVPASRGFCLTLRSSLEAFRAALRRNDITLPPEDL
Summary
Uniprot
H9J7U6
A0A2Z5UMI9
A0A2H1WL89
A0A212F1T2
S4PD68
A0A194QRY6
+ More
A0A2A4JGP9 A0A2A4JHK1 A0A194QB72 A0A1B6F3L3 A0A1B6HB59 A0A3R7SYG0 A0A0A9Z4M3 A0A182S688 A0A146KZ01 A0A088A5L9 A0A2A3E541 A0A0M8ZST2 A0A3R7MFN5 A0A310SVJ8 A0A0J7K8A6 Q177P2 A0A1S4FCP9 F4WGT7 E2A6R6 A0A0P4WW47 A0A158NFG9 A0A195BCH6 A0A151JUU8 E9ITG6 A0A2J7QWP0 A0A151WPH2 A0A232EP30 A0A0L7R2N9 A0A195D6X5 K7IZ35 A0A154PRB3 A0A023EYQ1 A0A026WKB5 A0A0P4WP78 A0A195DAM2 A0A0V0G6I3 A0A224X9X2 A0A069DZS3 A0A171ADL9 A0A1B6CR24 E2C4G2 A0A0K8VG06 A0A336MDJ5 A0A0P4VTQ4 A0A1Q3FIR0 A0A1Q3FIR6 A0A336LYQ9 A0A182XVP0 A0A182P300 B0W911 A0A182HEW6 T1P7J6 W8BCA6 E0VN24 A0A1J1HL30 A0A182IVH4 A0A034VER4 A0A0K8ULW2 A0A182LUL4 A0A2M4CSB7 A0A084W918 A0A1A9UVH6 A0A1B0FQR3 A0A182JWR4 A0A1A9ZZ93 A0A182W0D7 A0A182N6M8 T1JBM6 T1HCU1 A0A182F6H2 A0A182Q3T4 A0A2M4B9Y1 A0A1A9WYQ4 A0A2M4B9Q3 A0A1A9YT37 A0A1I8PX28 A0A224Z9G1 A0A131YLB0 A0A224ZBM3 A0A131YJS8 A0A182UZU2 Q7QBM6 A0A182TRJ6 A0A1B0B3E5 A0A182HIZ2 A0A182X8D8 A0A182LC45 A0A2P8YLV3 A0A087SWE1 A0A1I8NFF5 A0A2R5LIX0 A0A067QTW6 L7LVL6 A0A131XKB5
A0A2A4JGP9 A0A2A4JHK1 A0A194QB72 A0A1B6F3L3 A0A1B6HB59 A0A3R7SYG0 A0A0A9Z4M3 A0A182S688 A0A146KZ01 A0A088A5L9 A0A2A3E541 A0A0M8ZST2 A0A3R7MFN5 A0A310SVJ8 A0A0J7K8A6 Q177P2 A0A1S4FCP9 F4WGT7 E2A6R6 A0A0P4WW47 A0A158NFG9 A0A195BCH6 A0A151JUU8 E9ITG6 A0A2J7QWP0 A0A151WPH2 A0A232EP30 A0A0L7R2N9 A0A195D6X5 K7IZ35 A0A154PRB3 A0A023EYQ1 A0A026WKB5 A0A0P4WP78 A0A195DAM2 A0A0V0G6I3 A0A224X9X2 A0A069DZS3 A0A171ADL9 A0A1B6CR24 E2C4G2 A0A0K8VG06 A0A336MDJ5 A0A0P4VTQ4 A0A1Q3FIR0 A0A1Q3FIR6 A0A336LYQ9 A0A182XVP0 A0A182P300 B0W911 A0A182HEW6 T1P7J6 W8BCA6 E0VN24 A0A1J1HL30 A0A182IVH4 A0A034VER4 A0A0K8ULW2 A0A182LUL4 A0A2M4CSB7 A0A084W918 A0A1A9UVH6 A0A1B0FQR3 A0A182JWR4 A0A1A9ZZ93 A0A182W0D7 A0A182N6M8 T1JBM6 T1HCU1 A0A182F6H2 A0A182Q3T4 A0A2M4B9Y1 A0A1A9WYQ4 A0A2M4B9Q3 A0A1A9YT37 A0A1I8PX28 A0A224Z9G1 A0A131YLB0 A0A224ZBM3 A0A131YJS8 A0A182UZU2 Q7QBM6 A0A182TRJ6 A0A1B0B3E5 A0A182HIZ2 A0A182X8D8 A0A182LC45 A0A2P8YLV3 A0A087SWE1 A0A1I8NFF5 A0A2R5LIX0 A0A067QTW6 L7LVL6 A0A131XKB5
Pubmed
19121390
22118469
23622113
26354079
25401762
26823975
+ More
17510324 21719571 20798317 21347285 21282665 28648823 20075255 25474469 24508170 30249741 26334808 27129103 25244985 26483478 24495485 20566863 25348373 24438588 28797301 26830274 12364791 14747013 17210077 20966253 29403074 25315136 24845553 25576852 28049606
17510324 21719571 20798317 21347285 21282665 28648823 20075255 25474469 24508170 30249741 26334808 27129103 25244985 26483478 24495485 20566863 25348373 24438588 28797301 26830274 12364791 14747013 17210077 20966253 29403074 25315136 24845553 25576852 28049606
EMBL
BABH01020117
BABH01020118
AP017548
BBB06957.1
ODYU01009425
SOQ53843.1
+ More
AGBW02010829 OWR47674.1 GAIX01005072 JAA87488.1 KQ461155 KPJ08252.1 NWSH01001429 PCG71255.1 PCG71256.1 KQ459232 KPJ02679.1 GECZ01024893 GECZ01016377 JAS44876.1 JAS53392.1 GECU01035770 JAS71936.1 QCYY01000991 ROT81312.1 GBHO01023831 GBHO01023829 GBHO01023828 GBHO01023826 GBHO01023825 GBHO01023824 GBHO01006853 GBHO01006849 GBRD01014963 GBRD01014962 JAG19773.1 JAG19775.1 JAG19776.1 JAG19778.1 JAG19779.1 JAG19780.1 JAG36751.1 JAG36755.1 JAG50863.1 GDHC01017231 JAQ01398.1 KZ288400 PBC26289.1 KQ435912 KOX68893.1 QCYY01000992 ROT81303.1 KQ760116 OAD61943.1 LBMM01012045 KMQ86451.1 CH477373 EAT42376.1 GL888147 EGI66556.1 GL437197 EFN70860.1 GDRN01055014 JAI66025.1 ADTU01014480 KQ976514 KYM82266.1 KQ981724 KYN37239.1 GL765555 EFZ16116.1 NEVH01009422 PNF33000.1 KQ982877 KYQ49701.1 NNAY01003020 OXU20104.1 KQ414665 KOC65142.1 KQ976750 KYN08640.1 KQ435056 KZC14287.1 GBBI01004419 JAC14293.1 KK107167 QOIP01000010 EZA56492.1 RLU17790.1 GDRN01055013 JAI66026.1 KQ981063 KYN09960.1 GECL01002768 JAP03356.1 GFTR01008722 JAW07704.1 GBGD01000140 JAC88749.1 GEMB01001195 JAS01953.1 GEDC01021378 GEDC01004218 JAS15920.1 JAS33080.1 GL452471 EFN77146.1 GDHF01014527 JAI37787.1 UFQS01001009 UFQT01001009 SSX08556.1 SSX28472.1 GDKW01002309 JAI54286.1 GFDL01007578 JAV27467.1 GFDL01007586 JAV27459.1 UFQT01000234 SSX22151.1 DS231861 EDS39449.1 JXUM01132778 KQ567891 KXJ69233.1 KA644439 AFP59068.1 GAMC01015764 JAB90791.1 DS235332 EEB14790.1 CVRI01000010 CRK88767.1 GAKP01018707 JAC40245.1 GDHF01024615 JAI27699.1 AXCM01000558 GGFL01004025 MBW68203.1 ATLV01021572 KE525320 KFB46712.1 CCAG010018120 JH432011 ACPB03000177 AXCN02000299 GGFJ01000660 MBW49801.1 GGFJ01000615 MBW49756.1 GFPF01013135 MAA24281.1 GEDV01009856 JAP78701.1 GFPF01013136 MAA24282.1 GEDV01009857 JAP78700.1 AAAB01008879 EAA08452.5 EGK96947.1 JXJN01007813 APCN01002105 PYGN01000503 PSN45221.1 KK112250 KFM57180.1 GGLE01005249 MBY09375.1 KK853278 KDR09067.1 GACK01010091 JAA54943.1 GEFH01001052 JAP67529.1
AGBW02010829 OWR47674.1 GAIX01005072 JAA87488.1 KQ461155 KPJ08252.1 NWSH01001429 PCG71255.1 PCG71256.1 KQ459232 KPJ02679.1 GECZ01024893 GECZ01016377 JAS44876.1 JAS53392.1 GECU01035770 JAS71936.1 QCYY01000991 ROT81312.1 GBHO01023831 GBHO01023829 GBHO01023828 GBHO01023826 GBHO01023825 GBHO01023824 GBHO01006853 GBHO01006849 GBRD01014963 GBRD01014962 JAG19773.1 JAG19775.1 JAG19776.1 JAG19778.1 JAG19779.1 JAG19780.1 JAG36751.1 JAG36755.1 JAG50863.1 GDHC01017231 JAQ01398.1 KZ288400 PBC26289.1 KQ435912 KOX68893.1 QCYY01000992 ROT81303.1 KQ760116 OAD61943.1 LBMM01012045 KMQ86451.1 CH477373 EAT42376.1 GL888147 EGI66556.1 GL437197 EFN70860.1 GDRN01055014 JAI66025.1 ADTU01014480 KQ976514 KYM82266.1 KQ981724 KYN37239.1 GL765555 EFZ16116.1 NEVH01009422 PNF33000.1 KQ982877 KYQ49701.1 NNAY01003020 OXU20104.1 KQ414665 KOC65142.1 KQ976750 KYN08640.1 KQ435056 KZC14287.1 GBBI01004419 JAC14293.1 KK107167 QOIP01000010 EZA56492.1 RLU17790.1 GDRN01055013 JAI66026.1 KQ981063 KYN09960.1 GECL01002768 JAP03356.1 GFTR01008722 JAW07704.1 GBGD01000140 JAC88749.1 GEMB01001195 JAS01953.1 GEDC01021378 GEDC01004218 JAS15920.1 JAS33080.1 GL452471 EFN77146.1 GDHF01014527 JAI37787.1 UFQS01001009 UFQT01001009 SSX08556.1 SSX28472.1 GDKW01002309 JAI54286.1 GFDL01007578 JAV27467.1 GFDL01007586 JAV27459.1 UFQT01000234 SSX22151.1 DS231861 EDS39449.1 JXUM01132778 KQ567891 KXJ69233.1 KA644439 AFP59068.1 GAMC01015764 JAB90791.1 DS235332 EEB14790.1 CVRI01000010 CRK88767.1 GAKP01018707 JAC40245.1 GDHF01024615 JAI27699.1 AXCM01000558 GGFL01004025 MBW68203.1 ATLV01021572 KE525320 KFB46712.1 CCAG010018120 JH432011 ACPB03000177 AXCN02000299 GGFJ01000660 MBW49801.1 GGFJ01000615 MBW49756.1 GFPF01013135 MAA24281.1 GEDV01009856 JAP78701.1 GFPF01013136 MAA24282.1 GEDV01009857 JAP78700.1 AAAB01008879 EAA08452.5 EGK96947.1 JXJN01007813 APCN01002105 PYGN01000503 PSN45221.1 KK112250 KFM57180.1 GGLE01005249 MBY09375.1 KK853278 KDR09067.1 GACK01010091 JAA54943.1 GEFH01001052 JAP67529.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000218220
UP000053268
UP000283509
+ More
UP000075901 UP000005203 UP000242457 UP000053105 UP000036403 UP000008820 UP000007755 UP000000311 UP000005205 UP000078540 UP000078541 UP000235965 UP000075809 UP000215335 UP000053825 UP000078542 UP000002358 UP000076502 UP000053097 UP000279307 UP000078492 UP000008237 UP000076408 UP000075885 UP000002320 UP000069940 UP000249989 UP000009046 UP000183832 UP000075880 UP000075883 UP000030765 UP000078200 UP000092444 UP000075881 UP000092445 UP000075920 UP000075884 UP000015103 UP000069272 UP000075886 UP000091820 UP000092443 UP000095300 UP000075903 UP000007062 UP000075902 UP000092460 UP000075840 UP000076407 UP000075882 UP000245037 UP000054359 UP000095301 UP000027135
UP000075901 UP000005203 UP000242457 UP000053105 UP000036403 UP000008820 UP000007755 UP000000311 UP000005205 UP000078540 UP000078541 UP000235965 UP000075809 UP000215335 UP000053825 UP000078542 UP000002358 UP000076502 UP000053097 UP000279307 UP000078492 UP000008237 UP000076408 UP000075885 UP000002320 UP000069940 UP000249989 UP000009046 UP000183832 UP000075880 UP000075883 UP000030765 UP000078200 UP000092444 UP000075881 UP000092445 UP000075920 UP000075884 UP000015103 UP000069272 UP000075886 UP000091820 UP000092443 UP000095300 UP000075903 UP000007062 UP000075902 UP000092460 UP000075840 UP000076407 UP000075882 UP000245037 UP000054359 UP000095301 UP000027135
PRIDE
Pfam
Interpro
IPR016135
UBQ-conjugating_enzyme/RWD
+ More
IPR000608 UBQ-conjugat_E2
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR000629 RNA-helicase_DEAD-box_CS
IPR036318 FAD-bd_PCMH-like_sf
IPR004113 FAD-linked_oxidase_C
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016171 Vanillyl_alc_oxidase_C-sub2
IPR006094 Oxid_FAD_bind_N
IPR000608 UBQ-conjugat_E2
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR027417 P-loop_NTPase
IPR011545 DEAD/DEAH_box_helicase_dom
IPR014014 RNA_helicase_DEAD_Q_motif
IPR000629 RNA-helicase_DEAD-box_CS
IPR036318 FAD-bd_PCMH-like_sf
IPR004113 FAD-linked_oxidase_C
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR016171 Vanillyl_alc_oxidase_C-sub2
IPR006094 Oxid_FAD_bind_N
Gene 3D
ProteinModelPortal
H9J7U6
A0A2Z5UMI9
A0A2H1WL89
A0A212F1T2
S4PD68
A0A194QRY6
+ More
A0A2A4JGP9 A0A2A4JHK1 A0A194QB72 A0A1B6F3L3 A0A1B6HB59 A0A3R7SYG0 A0A0A9Z4M3 A0A182S688 A0A146KZ01 A0A088A5L9 A0A2A3E541 A0A0M8ZST2 A0A3R7MFN5 A0A310SVJ8 A0A0J7K8A6 Q177P2 A0A1S4FCP9 F4WGT7 E2A6R6 A0A0P4WW47 A0A158NFG9 A0A195BCH6 A0A151JUU8 E9ITG6 A0A2J7QWP0 A0A151WPH2 A0A232EP30 A0A0L7R2N9 A0A195D6X5 K7IZ35 A0A154PRB3 A0A023EYQ1 A0A026WKB5 A0A0P4WP78 A0A195DAM2 A0A0V0G6I3 A0A224X9X2 A0A069DZS3 A0A171ADL9 A0A1B6CR24 E2C4G2 A0A0K8VG06 A0A336MDJ5 A0A0P4VTQ4 A0A1Q3FIR0 A0A1Q3FIR6 A0A336LYQ9 A0A182XVP0 A0A182P300 B0W911 A0A182HEW6 T1P7J6 W8BCA6 E0VN24 A0A1J1HL30 A0A182IVH4 A0A034VER4 A0A0K8ULW2 A0A182LUL4 A0A2M4CSB7 A0A084W918 A0A1A9UVH6 A0A1B0FQR3 A0A182JWR4 A0A1A9ZZ93 A0A182W0D7 A0A182N6M8 T1JBM6 T1HCU1 A0A182F6H2 A0A182Q3T4 A0A2M4B9Y1 A0A1A9WYQ4 A0A2M4B9Q3 A0A1A9YT37 A0A1I8PX28 A0A224Z9G1 A0A131YLB0 A0A224ZBM3 A0A131YJS8 A0A182UZU2 Q7QBM6 A0A182TRJ6 A0A1B0B3E5 A0A182HIZ2 A0A182X8D8 A0A182LC45 A0A2P8YLV3 A0A087SWE1 A0A1I8NFF5 A0A2R5LIX0 A0A067QTW6 L7LVL6 A0A131XKB5
A0A2A4JGP9 A0A2A4JHK1 A0A194QB72 A0A1B6F3L3 A0A1B6HB59 A0A3R7SYG0 A0A0A9Z4M3 A0A182S688 A0A146KZ01 A0A088A5L9 A0A2A3E541 A0A0M8ZST2 A0A3R7MFN5 A0A310SVJ8 A0A0J7K8A6 Q177P2 A0A1S4FCP9 F4WGT7 E2A6R6 A0A0P4WW47 A0A158NFG9 A0A195BCH6 A0A151JUU8 E9ITG6 A0A2J7QWP0 A0A151WPH2 A0A232EP30 A0A0L7R2N9 A0A195D6X5 K7IZ35 A0A154PRB3 A0A023EYQ1 A0A026WKB5 A0A0P4WP78 A0A195DAM2 A0A0V0G6I3 A0A224X9X2 A0A069DZS3 A0A171ADL9 A0A1B6CR24 E2C4G2 A0A0K8VG06 A0A336MDJ5 A0A0P4VTQ4 A0A1Q3FIR0 A0A1Q3FIR6 A0A336LYQ9 A0A182XVP0 A0A182P300 B0W911 A0A182HEW6 T1P7J6 W8BCA6 E0VN24 A0A1J1HL30 A0A182IVH4 A0A034VER4 A0A0K8ULW2 A0A182LUL4 A0A2M4CSB7 A0A084W918 A0A1A9UVH6 A0A1B0FQR3 A0A182JWR4 A0A1A9ZZ93 A0A182W0D7 A0A182N6M8 T1JBM6 T1HCU1 A0A182F6H2 A0A182Q3T4 A0A2M4B9Y1 A0A1A9WYQ4 A0A2M4B9Q3 A0A1A9YT37 A0A1I8PX28 A0A224Z9G1 A0A131YLB0 A0A224ZBM3 A0A131YJS8 A0A182UZU2 Q7QBM6 A0A182TRJ6 A0A1B0B3E5 A0A182HIZ2 A0A182X8D8 A0A182LC45 A0A2P8YLV3 A0A087SWE1 A0A1I8NFF5 A0A2R5LIX0 A0A067QTW6 L7LVL6 A0A131XKB5
PDB
3CEG
E-value=3.66729e-36,
Score=378
Ontologies
GO
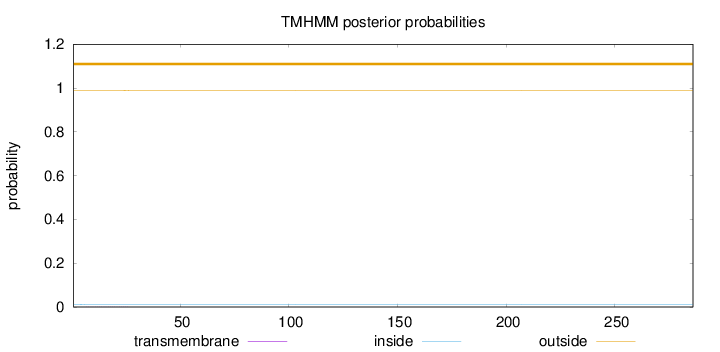
Topology
Length:
286
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01926
Exp number, first 60 AAs:
0.01681
Total prob of N-in:
0.01089
outside
1 - 286
Population Genetic Test Statistics
Pi
170.052937
Theta
131.062169
Tajima's D
0.862853
CLR
0
CSRT
0.619119044047798
Interpretation
Uncertain
