Pre Gene Modal
BGIBMGA005584
Annotation
PREDICTED:_Usher_syndrome_type-1G_protein_homolog_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.674
Sequence
CDS
ATGTCCACGGACAGGTTCCACAAAGCAGCCAAAGATGGTCTACTGGAGGTACTGCGGGAAGCGACACGCAAAGAATGCAATAATAAAGATGAATCTGGAATGACTCCCACGCTGTGGGCTGCTTTCGAAGGACACATAGAAGCCCTGAGACTACTATGCGGGCGAGGCGGTGAACCAGACAAGGCCGACTATTTCGGCAATACAGCCCTTCATCTAGCAGCGGCCCGGGGTCACAAGGAATGCGTGACTTTCCTGGTCAACTTCGGAGCTAATGTGTACGCTATGGACGTCGACGGTCACTCCGCTCAGGAGCTGGCCGCTATTAATGGCAGAGAGGATATACTCCGGTACCTGGACCAGTCTACAGCAAAACTAGAAAACAATGATAAGAAGAAATCGAAAGCGCTCAAAGAGAAAGCCAGAAAAGACTACGAGAAATCGCAGAAGCAGTATCAGAAAAGACAGAGCAAAGCTGATCTCATGGCCGAGAAGGAAATGAAGAAGCTGGCCAAAGAGTGGGAACAGGGTTATGGTGAGGAGGTGTCCACTATGCCACATCGGCCCAGCAACGTGCTAATAGCGCTCAAGCAAAAGATGATCAGATCTACAAGTCAGGGTAACCTCCTGGCGGACGAGCAGCCCCGCCCGACGTACAGCACGCTGGTCGGCACCGTGAACTCGGGCGTCCGGGGCCGCGGCGCCGTCTACAAGAAGGCCCTCGCCAGCAAACTGCGCAACAACACGATCGGCCGTAATGGACCGAATGATGACTTCAAGGTTGGCGAATTAGAAACGACCGGTCGTCGTTCTCTGACGTCATTATCCGGGCTCCGTCGCGACAGTGAAGTGATGTACGTGGGCACGTTGGGCGCGGGACCGCAACACCGGGGGAACGTTACCGACGTGTTCACGGAGGACACCGATCAGAAGAAACCGCCACTGACAAGATCGGCAAGTCAACCGGATTTCACGGTTCTGTCTGGAGAAGACAGCGGCATAGGACAGGAGGTGATGCTGCAAGAGCCCGCCAGTATATTCGACAGACCAGGATTCGGGAGTGTAGCCTTTAGACGGTCCATAACGGCCACGCTGGGCGCGATGCCGGGCGCGCCGGGCGAGGACCTGTCCATCGGCTCGGCGGGGTCACTGGCCGCGCGCCACGCCCCCCGCACCGCCTACCGCCCCGCCGAGTGGACCGAGCCGCACTCAGAGAGCTCGACGATAACTTCGGACGAGGAGGGGGAGGCCGATGAGTCCGGCTCGGCGTCCCTAGAGCGTTTCCTGACCGCGTGGGGCCTCTCGCAGTACGTGCAGAAGTTTCGAGACGAACAAATCGACTTGGAAGCGCTCATGCTGCTGACCGAGAGCGACATGAAATCGCTCGGCCTACCTCTAGGTCCGTACAGAAAACTCGTCACAGCTGTCCAAGAGAGGAAACAGGCGTTAACTAACCCGGGACCCATAGTCGACACGGCGATCTGA
Protein
MSTDRFHKAAKDGLLEVLREATRKECNNKDESGMTPTLWAAFEGHIEALRLLCGRGGEPDKADYFGNTALHLAAARGHKECVTFLVNFGANVYAMDVDGHSAQELAAINGREDILRYLDQSTAKLENNDKKKSKALKEKARKDYEKSQKQYQKRQSKADLMAEKEMKKLAKEWEQGYGEEVSTMPHRPSNVLIALKQKMIRSTSQGNLLADEQPRPTYSTLVGTVNSGVRGRGAVYKKALASKLRNNTIGRNGPNDDFKVGELETTGRRSLTSLSGLRRDSEVMYVGTLGAGPQHRGNVTDVFTEDTDQKKPPLTRSASQPDFTVLSGEDSGIGQEVMLQEPASIFDRPGFGSVAFRRSITATLGAMPGAPGEDLSIGSAGSLAARHAPRTAYRPAEWTEPHSESSTITSDEEGEADESGSASLERFLTAWGLSQYVQKFRDEQIDLEALMLLTESDMKSLGLPLGPYRKLVTAVQERKQALTNPGPIVDTAI
Summary
Uniprot
H9J7U2
A0A2A4JZF6
A0A194QXF9
A0A212EPI5
A0A2H1V7B6
A0A2H1WCR0
+ More
A0A2H1WCZ4 A0A2H1V616 A0A0L7L369 A0A2J7QN27 A0A1W4X2C7 A0A1Y1LJQ2 A0A1Y1LI48 A0A182PQN1 A0A182L3D5 A0A182I6Y9 Q7QIQ9 A0A182WWQ3 A0A182TE61 A0A182VLE8 A0A182JMA4 A0A182JUT1 A0A1Q3FA49 W5J9R6 A0A2M4ATA1 A0A182NJL9 W8BFN7 A0A182FDS6 B0XBG5 A0A0K8W307 A0A182R336 A0A182Y294 A0A034VSM7 A0A0M4EFX5 T1DN71 E2ASM7 N6SSA6 A0A0A1WL69 A0A3B0JNQ5 J3JVH7 Q16T60 F4X1M1 A0A158NKJ3 A0A087ZTW2 B4MJQ2 B3MDC4 A0A0L0C802 B4GH88 Q28XZ3 A0A182G121 A0A3L8E1I3 A0A1W4VCP6 A0A1I8P026 A0A182QTZ5 Q7JQL2 B4LM39 A0A1J1I7Z9 A0A0J9RBH0 B3NRT0 B4P4W5 A0A151WT62 B4HPY8 A0A336MMG4 A0A195EK62 A0A2A3EEC3 A0A0P4VQA1 A0A1L8DSV1 A0A182WL37 A0A195D031 A0A1A9X3Q3 A0A182MS25 A0A154PMS9 A0A026WVB3 A0A067QQN4 A0A1B0BVB0 A0A1B6CD13 A0A0C9QN17 D6WX01 A0A310SPV3 A0A1D2N385 W8B5C3 A0A1B6GQ44 A0A0P5J4J4 A0A0P5J835 E9FXB7 A0A0P5J422 A0A0P6HP02 A0A0P5UID4 A0A0P6AFN4 T1JIG7 A0A0L7QR70 A0A1B6KI22
A0A2H1WCZ4 A0A2H1V616 A0A0L7L369 A0A2J7QN27 A0A1W4X2C7 A0A1Y1LJQ2 A0A1Y1LI48 A0A182PQN1 A0A182L3D5 A0A182I6Y9 Q7QIQ9 A0A182WWQ3 A0A182TE61 A0A182VLE8 A0A182JMA4 A0A182JUT1 A0A1Q3FA49 W5J9R6 A0A2M4ATA1 A0A182NJL9 W8BFN7 A0A182FDS6 B0XBG5 A0A0K8W307 A0A182R336 A0A182Y294 A0A034VSM7 A0A0M4EFX5 T1DN71 E2ASM7 N6SSA6 A0A0A1WL69 A0A3B0JNQ5 J3JVH7 Q16T60 F4X1M1 A0A158NKJ3 A0A087ZTW2 B4MJQ2 B3MDC4 A0A0L0C802 B4GH88 Q28XZ3 A0A182G121 A0A3L8E1I3 A0A1W4VCP6 A0A1I8P026 A0A182QTZ5 Q7JQL2 B4LM39 A0A1J1I7Z9 A0A0J9RBH0 B3NRT0 B4P4W5 A0A151WT62 B4HPY8 A0A336MMG4 A0A195EK62 A0A2A3EEC3 A0A0P4VQA1 A0A1L8DSV1 A0A182WL37 A0A195D031 A0A1A9X3Q3 A0A182MS25 A0A154PMS9 A0A026WVB3 A0A067QQN4 A0A1B0BVB0 A0A1B6CD13 A0A0C9QN17 D6WX01 A0A310SPV3 A0A1D2N385 W8B5C3 A0A1B6GQ44 A0A0P5J4J4 A0A0P5J835 E9FXB7 A0A0P5J422 A0A0P6HP02 A0A0P5UID4 A0A0P6AFN4 T1JIG7 A0A0L7QR70 A0A1B6KI22
Pubmed
19121390
26354079
22118469
26227816
28004739
20966253
+ More
12364791 14747013 17210077 20920257 23761445 24495485 25244985 25348373 20798317 23537049 25830018 22516182 17510324 21719571 21347285 17994087 26108605 15632085 23185243 26483478 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18057021 17550304 24508170 24845553 18362917 19820115 27289101 21292972
12364791 14747013 17210077 20920257 23761445 24495485 25244985 25348373 20798317 23537049 25830018 22516182 17510324 21719571 21347285 17994087 26108605 15632085 23185243 26483478 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18057021 17550304 24508170 24845553 18362917 19820115 27289101 21292972
EMBL
BABH01020122
BABH01020123
NWSH01000358
PCG77024.1
KQ461155
KPJ08261.1
+ More
AGBW02013457 OWR43384.1 ODYU01000866 SOQ36272.1 ODYU01007772 SOQ50827.1 SOQ50826.1 SOQ36271.1 JTDY01003256 KOB69865.1 NEVH01013201 PNF29990.1 GEZM01060142 JAV71247.1 GEZM01060143 JAV71246.1 APCN01003712 AAAB01008807 EAA04767.4 GFDL01010646 JAV24399.1 ADMH02001746 ETN61207.1 GGFK01010673 MBW43994.1 GAMC01018076 JAB88479.1 DS232633 EDS44277.1 GDHF01007059 JAI45255.1 GAKP01013840 JAC45112.1 CP012524 ALC42338.1 GAMD01003135 JAA98455.1 GL442333 EFN63562.1 APGK01058608 APGK01058609 KB741291 KB632343 ENN70529.1 ERL93027.1 GBXI01014528 JAC99763.1 OUUW01000001 SPP74281.1 BT127245 AEE62207.1 CH477659 EAT37647.1 GL888531 EGI59679.1 ADTU01002070 CH963846 EDW72341.1 CH902619 EDV36372.1 JRES01000780 KNC28381.1 CH479183 EDW35858.1 CM000071 EAL26173.1 KRT02898.1 JXUM01038027 KQ561151 KXJ79547.1 QOIP01000001 RLU26527.1 AXCN02000438 AE013599 BT009925 AAF58438.1 AAO41397.1 AAQ21582.1 CH940648 EDW59959.2 CVRI01000043 CRK96405.1 CM002911 KMY93378.1 KMY93379.1 CH954179 EDV56232.1 KQS62777.1 CM000158 EDW90686.1 KQ982757 KYQ51102.1 CH480816 EDW47651.1 UFQT01000784 SSX27198.1 KQ978801 KYN28307.1 KZ288268 PBC30060.1 GDRN01108547 JAI57214.1 GFDF01004532 JAV09552.1 KQ977110 KYN05734.1 AXCM01004683 KQ434973 KZC12764.1 KK107087 EZA59990.1 KK853541 KDR06891.1 JXJN01021173 GEDC01026053 JAS11245.1 GBYB01002052 JAG71819.1 KQ971361 EFA08068.1 KQ761834 OAD56697.1 LJIJ01000259 ODM99728.1 GAMC01018074 JAB88481.1 GECZ01005215 JAS64554.1 GDIQ01206719 JAK45006.1 GDIQ01205265 JAK46460.1 GL732526 EFX88296.1 GDIQ01203502 JAK48223.1 GDIQ01024259 JAN70478.1 GDIP01116636 JAL87078.1 GDIP01031346 JAM72369.1 JH431789 KQ414784 KOC61059.1 GEBQ01028869 JAT11108.1
AGBW02013457 OWR43384.1 ODYU01000866 SOQ36272.1 ODYU01007772 SOQ50827.1 SOQ50826.1 SOQ36271.1 JTDY01003256 KOB69865.1 NEVH01013201 PNF29990.1 GEZM01060142 JAV71247.1 GEZM01060143 JAV71246.1 APCN01003712 AAAB01008807 EAA04767.4 GFDL01010646 JAV24399.1 ADMH02001746 ETN61207.1 GGFK01010673 MBW43994.1 GAMC01018076 JAB88479.1 DS232633 EDS44277.1 GDHF01007059 JAI45255.1 GAKP01013840 JAC45112.1 CP012524 ALC42338.1 GAMD01003135 JAA98455.1 GL442333 EFN63562.1 APGK01058608 APGK01058609 KB741291 KB632343 ENN70529.1 ERL93027.1 GBXI01014528 JAC99763.1 OUUW01000001 SPP74281.1 BT127245 AEE62207.1 CH477659 EAT37647.1 GL888531 EGI59679.1 ADTU01002070 CH963846 EDW72341.1 CH902619 EDV36372.1 JRES01000780 KNC28381.1 CH479183 EDW35858.1 CM000071 EAL26173.1 KRT02898.1 JXUM01038027 KQ561151 KXJ79547.1 QOIP01000001 RLU26527.1 AXCN02000438 AE013599 BT009925 AAF58438.1 AAO41397.1 AAQ21582.1 CH940648 EDW59959.2 CVRI01000043 CRK96405.1 CM002911 KMY93378.1 KMY93379.1 CH954179 EDV56232.1 KQS62777.1 CM000158 EDW90686.1 KQ982757 KYQ51102.1 CH480816 EDW47651.1 UFQT01000784 SSX27198.1 KQ978801 KYN28307.1 KZ288268 PBC30060.1 GDRN01108547 JAI57214.1 GFDF01004532 JAV09552.1 KQ977110 KYN05734.1 AXCM01004683 KQ434973 KZC12764.1 KK107087 EZA59990.1 KK853541 KDR06891.1 JXJN01021173 GEDC01026053 JAS11245.1 GBYB01002052 JAG71819.1 KQ971361 EFA08068.1 KQ761834 OAD56697.1 LJIJ01000259 ODM99728.1 GAMC01018074 JAB88481.1 GECZ01005215 JAS64554.1 GDIQ01206719 JAK45006.1 GDIQ01205265 JAK46460.1 GL732526 EFX88296.1 GDIQ01203502 JAK48223.1 GDIQ01024259 JAN70478.1 GDIP01116636 JAL87078.1 GDIP01031346 JAM72369.1 JH431789 KQ414784 KOC61059.1 GEBQ01028869 JAT11108.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000037510
UP000235965
+ More
UP000192223 UP000075885 UP000075882 UP000075840 UP000007062 UP000076407 UP000075902 UP000075903 UP000075880 UP000075881 UP000000673 UP000075884 UP000069272 UP000002320 UP000075900 UP000076408 UP000092553 UP000000311 UP000019118 UP000030742 UP000268350 UP000008820 UP000007755 UP000005205 UP000005203 UP000007798 UP000007801 UP000037069 UP000008744 UP000001819 UP000069940 UP000249989 UP000279307 UP000192221 UP000095300 UP000075886 UP000000803 UP000008792 UP000183832 UP000008711 UP000002282 UP000075809 UP000001292 UP000078492 UP000242457 UP000075920 UP000078542 UP000091820 UP000075883 UP000076502 UP000053097 UP000027135 UP000092460 UP000007266 UP000094527 UP000000305 UP000053825
UP000192223 UP000075885 UP000075882 UP000075840 UP000007062 UP000076407 UP000075902 UP000075903 UP000075880 UP000075881 UP000000673 UP000075884 UP000069272 UP000002320 UP000075900 UP000076408 UP000092553 UP000000311 UP000019118 UP000030742 UP000268350 UP000008820 UP000007755 UP000005205 UP000005203 UP000007798 UP000007801 UP000037069 UP000008744 UP000001819 UP000069940 UP000249989 UP000279307 UP000192221 UP000095300 UP000075886 UP000000803 UP000008792 UP000183832 UP000008711 UP000002282 UP000075809 UP000001292 UP000078492 UP000242457 UP000075920 UP000078542 UP000091820 UP000075883 UP000076502 UP000053097 UP000027135 UP000092460 UP000007266 UP000094527 UP000000305 UP000053825
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J7U2
A0A2A4JZF6
A0A194QXF9
A0A212EPI5
A0A2H1V7B6
A0A2H1WCR0
+ More
A0A2H1WCZ4 A0A2H1V616 A0A0L7L369 A0A2J7QN27 A0A1W4X2C7 A0A1Y1LJQ2 A0A1Y1LI48 A0A182PQN1 A0A182L3D5 A0A182I6Y9 Q7QIQ9 A0A182WWQ3 A0A182TE61 A0A182VLE8 A0A182JMA4 A0A182JUT1 A0A1Q3FA49 W5J9R6 A0A2M4ATA1 A0A182NJL9 W8BFN7 A0A182FDS6 B0XBG5 A0A0K8W307 A0A182R336 A0A182Y294 A0A034VSM7 A0A0M4EFX5 T1DN71 E2ASM7 N6SSA6 A0A0A1WL69 A0A3B0JNQ5 J3JVH7 Q16T60 F4X1M1 A0A158NKJ3 A0A087ZTW2 B4MJQ2 B3MDC4 A0A0L0C802 B4GH88 Q28XZ3 A0A182G121 A0A3L8E1I3 A0A1W4VCP6 A0A1I8P026 A0A182QTZ5 Q7JQL2 B4LM39 A0A1J1I7Z9 A0A0J9RBH0 B3NRT0 B4P4W5 A0A151WT62 B4HPY8 A0A336MMG4 A0A195EK62 A0A2A3EEC3 A0A0P4VQA1 A0A1L8DSV1 A0A182WL37 A0A195D031 A0A1A9X3Q3 A0A182MS25 A0A154PMS9 A0A026WVB3 A0A067QQN4 A0A1B0BVB0 A0A1B6CD13 A0A0C9QN17 D6WX01 A0A310SPV3 A0A1D2N385 W8B5C3 A0A1B6GQ44 A0A0P5J4J4 A0A0P5J835 E9FXB7 A0A0P5J422 A0A0P6HP02 A0A0P5UID4 A0A0P6AFN4 T1JIG7 A0A0L7QR70 A0A1B6KI22
A0A2H1WCZ4 A0A2H1V616 A0A0L7L369 A0A2J7QN27 A0A1W4X2C7 A0A1Y1LJQ2 A0A1Y1LI48 A0A182PQN1 A0A182L3D5 A0A182I6Y9 Q7QIQ9 A0A182WWQ3 A0A182TE61 A0A182VLE8 A0A182JMA4 A0A182JUT1 A0A1Q3FA49 W5J9R6 A0A2M4ATA1 A0A182NJL9 W8BFN7 A0A182FDS6 B0XBG5 A0A0K8W307 A0A182R336 A0A182Y294 A0A034VSM7 A0A0M4EFX5 T1DN71 E2ASM7 N6SSA6 A0A0A1WL69 A0A3B0JNQ5 J3JVH7 Q16T60 F4X1M1 A0A158NKJ3 A0A087ZTW2 B4MJQ2 B3MDC4 A0A0L0C802 B4GH88 Q28XZ3 A0A182G121 A0A3L8E1I3 A0A1W4VCP6 A0A1I8P026 A0A182QTZ5 Q7JQL2 B4LM39 A0A1J1I7Z9 A0A0J9RBH0 B3NRT0 B4P4W5 A0A151WT62 B4HPY8 A0A336MMG4 A0A195EK62 A0A2A3EEC3 A0A0P4VQA1 A0A1L8DSV1 A0A182WL37 A0A195D031 A0A1A9X3Q3 A0A182MS25 A0A154PMS9 A0A026WVB3 A0A067QQN4 A0A1B0BVB0 A0A1B6CD13 A0A0C9QN17 D6WX01 A0A310SPV3 A0A1D2N385 W8B5C3 A0A1B6GQ44 A0A0P5J4J4 A0A0P5J835 E9FXB7 A0A0P5J422 A0A0P6HP02 A0A0P5UID4 A0A0P6AFN4 T1JIG7 A0A0L7QR70 A0A1B6KI22
PDB
4HQD
E-value=2.85607e-15,
Score=201
Ontologies
KEGG
GO
PANTHER
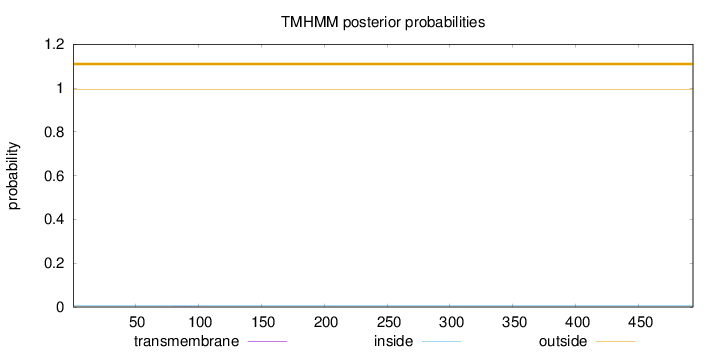
Topology
Length:
493
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00565
Exp number, first 60 AAs:
0.00035
Total prob of N-in:
0.00659
outside
1 - 493
Population Genetic Test Statistics
Pi
236.687698
Theta
174.383933
Tajima's D
1.600179
CLR
0.398372
CSRT
0.812959352032398
Interpretation
Uncertain
