Gene
KWMTBOMO10175 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013945
Annotation
hypothetical_protein_FF38_04907_[Lucilia_cuprina]
Location in the cell
Cytoplasmic Reliability : 3.574
Sequence
CDS
ATGTGCGACGAAGAAGTTGCCGCGTTGGTAGTAGACAATGGCTCCGGTATGTGCAAGGCCGGTTTCGCAGGAGATGATGCTCCTCGCGCCGTGTTCCCCTCGATCGTGGGAAGGCCCCGCCATCAGGGCGTGATGGTCGGTATGGGACAGAAGGACTCTTATGTAGGAGATGAGGCACAGAGCAAAAGAGGTATCCTGACCCTCAAATACCCCATCGAACACGGAATCGTCACTAACTGGGATGACATGGAGAAGATTTGGCATCATACCTTCTACAATGAGCTGCGTGTCGCCCCCGAGGAACACCCAGTCCTGCTCACTGAGGCTCCCCTCAACCCCAAGGCCAACAGAGAGAAGATGACCCAGATCATGTTCGAAACATTCAACACGCCCGCCATGTACGTCGCCATCCAAGCCGTGCTCTCGCTGTACGCGTCCGGTCGTACCACCGGTATCGTGCTGGACTCCGGCGACGGTGTCTCCCACACCGTGCCCATCTACGAGGGATACGCACTCCCCCACGCCATCCTGCGTCTGGACTTAGCCGGTCGCGACCTCACAGACTACCTCATGAAGATCCTCACCGAGCGCGGCTACTCGTTCACTACCACCGCCGAGCGGGAAATCGTTCGTGACATCAAGGAGAAGCTGTGCTACGTCGCTCTTGACTTCGAGCAGGAGATGGCCACCGCTGCATCCAGCAGCTCCCTCGAGAAGTCTTACGAACTTCCCGACGGTCAGGTCATCACCATCGGAAACGAAAGATTCCGTTGCCCAGAGGCTCTCTTCCAACCCTCGTTCTTGGGTATGGAAGCTTGCGGCATCCACGAGACCACATATAACTCCATCATGAAGTGCGACGTGGACATCCGTAAGGACTTGTACGCCAACACCGTATTGTCCGGTGGTACCACCATGTACCCTGGAATCGCCGACCGTATGCAAAAGGAAATCACAGCTCTCGCCCCATCGACAATGAAGATTAAGATCATTGCTCCTCCAGAGAGGAAGTACTCCGTATGGATCGGTGGATCGATCCTCGCCTCCCTCTCTACCTTCCAACAAATGTGGATCTCGAAACAAGAGTACGACGAGTCTGGCCCCTCCATTGTACACAGGAAGTGCTTCTAA
Protein
MCDEEVAALVVDNGSGMCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYVGDEAQSKRGILTLKYPIEHGIVTNWDDMEKIWHHTFYNELRVAPEEHPVLLTEAPLNPKANREKMTQIMFETFNTPAMYVAIQAVLSLYASGRTTGIVLDSGDGVSHTVPIYEGYALPHAILRLDLAGRDLTDYLMKILTERGYSFTTTAEREIVRDIKEKLCYVALDFEQEMATAASSSSLEKSYELPDGQVITIGNERFRCPEALFQPSFLGMEACGIHETTYNSIMKCDVDIRKDLYANTVLSGGTTMYPGIADRMQKEITALAPSTMKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQEYDESGPSIVHRKCF
Summary
Similarity
Belongs to the actin family.
Uniprot
A0A1B0G1D5
A0A1A9W215
A0A1A9X7U1
A0A1A9UUE4
A0A1L8E5F8
A0A1L8ECP3
+ More
A0A1L8E580 A0A0L0C8P9 A0A1L8ECD3 A0A323TKK8 B4R5A1 A0A2H4H004 A0A195CW50 A0A151JNS2 A0A059XKE4 A0A1W6EVQ4 D3TQK0 E9I8G6 A0A2A4JTM4 A0A154PID2 A0A0L7QNE1 I4DLZ5 A0A3S2M069 A0A1X9H806 A0A3B0JWK6 F4WEV7 T1E2Q6 T1DEW8 A0A140F3M8 A0A195B5V2 A0A1Q3EUM5 A0A1S6Q8B9 X2JCP8 B4H0C1 Q5XUC4 A0A0J7KZ38 R4FQ90 E3UKI0 A0A0M5J8B6 A0A2R4CIH4 A0A126CN36 A0A182YH06 F2ZBU0 A0A067RMN2 D8WKR5 A0A2H4ZTI5 I4DIX7 A0A0T6B3H8 A0A2M4CSZ9 A0A1B6F0W0 A0A151K130 D0PTT4 E2BJ05 A0A158NXD4 A0A140B7Z7 B4MTB7 A0A286NL51 B4I0H7 A0A310SF14 E2AFQ5 B4JM91 A0A2M3ZF62 A0A0G2YN85 A0A1A9Z4Q1 A0A2M3ZZM2 A0A3G1T170 A0A2P8Z2J3 A0A0K8TTW7 A0A212EPG4 A0A151X9U7 A0A1W4UVV0 S5M4E2 A0A2J7PLV1 A0A182LYK9 A0A182PUA7 A0A182R1F8 B4L1G6 A0A182T7I7 A0A125RA07 B3NSY7 A0A182UTZ1 Q29H46 G3FP77 A0A182N0N1 A0A182K3Z4 A0A0P8XGW8 B0WSZ2 A0A182WQH2 A0A1I8N9U1 A0A182U3B5 Q16QR7 B4MEM3 D6WVY4 A0A1B6CYP1 A0A182J2J4 A0A182X7E8 F5HJZ8 A0A1I8PU39 A0A182FZP4 B4Q1E2
A0A1L8E580 A0A0L0C8P9 A0A1L8ECD3 A0A323TKK8 B4R5A1 A0A2H4H004 A0A195CW50 A0A151JNS2 A0A059XKE4 A0A1W6EVQ4 D3TQK0 E9I8G6 A0A2A4JTM4 A0A154PID2 A0A0L7QNE1 I4DLZ5 A0A3S2M069 A0A1X9H806 A0A3B0JWK6 F4WEV7 T1E2Q6 T1DEW8 A0A140F3M8 A0A195B5V2 A0A1Q3EUM5 A0A1S6Q8B9 X2JCP8 B4H0C1 Q5XUC4 A0A0J7KZ38 R4FQ90 E3UKI0 A0A0M5J8B6 A0A2R4CIH4 A0A126CN36 A0A182YH06 F2ZBU0 A0A067RMN2 D8WKR5 A0A2H4ZTI5 I4DIX7 A0A0T6B3H8 A0A2M4CSZ9 A0A1B6F0W0 A0A151K130 D0PTT4 E2BJ05 A0A158NXD4 A0A140B7Z7 B4MTB7 A0A286NL51 B4I0H7 A0A310SF14 E2AFQ5 B4JM91 A0A2M3ZF62 A0A0G2YN85 A0A1A9Z4Q1 A0A2M3ZZM2 A0A3G1T170 A0A2P8Z2J3 A0A0K8TTW7 A0A212EPG4 A0A151X9U7 A0A1W4UVV0 S5M4E2 A0A2J7PLV1 A0A182LYK9 A0A182PUA7 A0A182R1F8 B4L1G6 A0A182T7I7 A0A125RA07 B3NSY7 A0A182UTZ1 Q29H46 G3FP77 A0A182N0N1 A0A182K3Z4 A0A0P8XGW8 B0WSZ2 A0A182WQH2 A0A1I8N9U1 A0A182U3B5 Q16QR7 B4MEM3 D6WVY4 A0A1B6CYP1 A0A182J2J4 A0A182X7E8 F5HJZ8 A0A1I8PU39 A0A182FZP4 B4Q1E2
Pubmed
26108605
17994087
24810421
20353571
21282665
22651552
+ More
21719571 24330624 26873291 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25244985 24845553 19757170 20798317 21347285 26930056 26346731 29403074 26369729 22118469 24551045 18057021 26826390 27142481 15632085 23185243 25315136 17510324 18362917 19820115 12364791 14747013 17210077 17550304
21719571 24330624 26873291 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25244985 24845553 19757170 20798317 21347285 26930056 26346731 29403074 26369729 22118469 24551045 18057021 26826390 27142481 15632085 23185243 25315136 17510324 18362917 19820115 12364791 14747013 17210077 17550304
EMBL
CCAG010000053
GFDF01000322
JAV13762.1
GFDG01002397
JAV16402.1
GFDF01000193
+ More
JAV13891.1 JRES01000755 KNC28651.1 GFDG01002398 JAV16401.1 MDVM02000069 PYZ99322.1 CM000366 EDX17197.1 KU884974 ARE68323.1 KQ977279 KYN04384.1 KQ978820 KYN28144.1 KJ612090 AIA25510.1 KY563399 ARK19808.1 EZ423702 ADD19978.1 GL761543 EFZ23137.1 NWSH01000606 PCG75361.1 KQ434924 KZC11593.1 KQ414855 KOC60133.1 AK402313 BAM18935.1 RSAL01000090 RVE48073.1 KX077710 ANA52570.1 OUUW01000003 SPP77736.1 GL888111 EGI67274.1 GALA01001080 JAA93772.1 GAMD01003356 JAA98234.1 KT819630 AML61188.1 KQ976595 KYM79637.1 GFDL01016025 JAV19020.1 KY499580 AQV11934.1 AE014298 AHN59386.1 CH479200 EDW29716.1 AY737530 AAU84923.1 LBMM01001888 KMQ95606.1 GAHY01000539 JAA76971.1 HM449854 ADO32989.1 CP012528 ALC49444.1 MG775274 AVR99436.1 KC702778 AHG54250.1 AB626808 BAK19478.1 KK852421 KDR24293.1 GU176616 ADJ67594.1 MF615617 AUG45771.1 AK401245 BAM17867.1 LJIG01016001 KRT81912.1 GGFL01004207 MBW68385.1 GECZ01025870 JAS43899.1 KQ981189 KYN45235.1 FJ196622 ACN32208.1 GL448543 EFN84350.1 ADTU01029809 KT860051 ALS03939.1 CH963851 EDW75356.1 KX809573 ASZ69794.1 CH480819 EDW53008.1 KQ779356 OAD51990.1 GL439124 EFN67734.1 CH916371 EDV91852.1 GGFM01006370 MBW27121.1 KP331524 AKI87970.1 GGFK01000634 MBW33955.1 MG846876 AXY94728.1 PYGN01000224 PSN50721.1 GDAI01000022 JAI17581.1 AGBW02013457 OWR43395.1 KQ982365 KYQ57141.1 KC879873 KC879874 KC879877 KC879890 KC879893 KC879907 KC879919 KC879920 KC879921 KC879928 KC879929 KC879931 KC879932 KC879945 KC879948 KC879957 KC879967 KC879968 AGR44785.1 AGR44786.1 AGR44789.1 AGR44802.1 AGR44805.1 AGR44819.1 AGR44831.1 AGR44832.1 AGR44833.1 AGR44840.1 AGR44841.1 AGR44843.1 AGR44844.1 AGR44857.1 AGR44860.1 AGR44869.1 AGR44879.1 AGR44880.1 NEVH01024423 PNF17320.1 AXCM01000748 AXCN02002234 CH933810 EDW06687.1 KRF93779.1 KU196666 KU365929 AMD16546.1 ANJ04645.1 CH954180 EDV45956.1 KQS29767.1 CH379064 EAL31912.1 KRT06670.1 JF751025 JF751026 AEO51758.1 AEO51759.1 CH902632 KPU74049.1 KPU74050.1 DS232078 EDS34125.1 CH477736 EAT36735.1 EAT36736.1 CH940664 EDW62998.1 KRF80869.1 KQ971359 EFA08633.1 GEDC01019627 GEDC01018696 GEDC01016947 JAS17671.1 JAS18602.1 JAS20351.1 AXCP01004891 AAAB01008847 EGK96552.1 EGK96553.1 CM000162 EDX02431.1 KRK06744.1
JAV13891.1 JRES01000755 KNC28651.1 GFDG01002398 JAV16401.1 MDVM02000069 PYZ99322.1 CM000366 EDX17197.1 KU884974 ARE68323.1 KQ977279 KYN04384.1 KQ978820 KYN28144.1 KJ612090 AIA25510.1 KY563399 ARK19808.1 EZ423702 ADD19978.1 GL761543 EFZ23137.1 NWSH01000606 PCG75361.1 KQ434924 KZC11593.1 KQ414855 KOC60133.1 AK402313 BAM18935.1 RSAL01000090 RVE48073.1 KX077710 ANA52570.1 OUUW01000003 SPP77736.1 GL888111 EGI67274.1 GALA01001080 JAA93772.1 GAMD01003356 JAA98234.1 KT819630 AML61188.1 KQ976595 KYM79637.1 GFDL01016025 JAV19020.1 KY499580 AQV11934.1 AE014298 AHN59386.1 CH479200 EDW29716.1 AY737530 AAU84923.1 LBMM01001888 KMQ95606.1 GAHY01000539 JAA76971.1 HM449854 ADO32989.1 CP012528 ALC49444.1 MG775274 AVR99436.1 KC702778 AHG54250.1 AB626808 BAK19478.1 KK852421 KDR24293.1 GU176616 ADJ67594.1 MF615617 AUG45771.1 AK401245 BAM17867.1 LJIG01016001 KRT81912.1 GGFL01004207 MBW68385.1 GECZ01025870 JAS43899.1 KQ981189 KYN45235.1 FJ196622 ACN32208.1 GL448543 EFN84350.1 ADTU01029809 KT860051 ALS03939.1 CH963851 EDW75356.1 KX809573 ASZ69794.1 CH480819 EDW53008.1 KQ779356 OAD51990.1 GL439124 EFN67734.1 CH916371 EDV91852.1 GGFM01006370 MBW27121.1 KP331524 AKI87970.1 GGFK01000634 MBW33955.1 MG846876 AXY94728.1 PYGN01000224 PSN50721.1 GDAI01000022 JAI17581.1 AGBW02013457 OWR43395.1 KQ982365 KYQ57141.1 KC879873 KC879874 KC879877 KC879890 KC879893 KC879907 KC879919 KC879920 KC879921 KC879928 KC879929 KC879931 KC879932 KC879945 KC879948 KC879957 KC879967 KC879968 AGR44785.1 AGR44786.1 AGR44789.1 AGR44802.1 AGR44805.1 AGR44819.1 AGR44831.1 AGR44832.1 AGR44833.1 AGR44840.1 AGR44841.1 AGR44843.1 AGR44844.1 AGR44857.1 AGR44860.1 AGR44869.1 AGR44879.1 AGR44880.1 NEVH01024423 PNF17320.1 AXCM01000748 AXCN02002234 CH933810 EDW06687.1 KRF93779.1 KU196666 KU365929 AMD16546.1 ANJ04645.1 CH954180 EDV45956.1 KQS29767.1 CH379064 EAL31912.1 KRT06670.1 JF751025 JF751026 AEO51758.1 AEO51759.1 CH902632 KPU74049.1 KPU74050.1 DS232078 EDS34125.1 CH477736 EAT36735.1 EAT36736.1 CH940664 EDW62998.1 KRF80869.1 KQ971359 EFA08633.1 GEDC01019627 GEDC01018696 GEDC01016947 JAS17671.1 JAS18602.1 JAS20351.1 AXCP01004891 AAAB01008847 EGK96552.1 EGK96553.1 CM000162 EDX02431.1 KRK06744.1
Proteomes
UP000092444
UP000091820
UP000092443
UP000078200
UP000037069
UP000247427
+ More
UP000000304 UP000078542 UP000078492 UP000218220 UP000076502 UP000053825 UP000283053 UP000268350 UP000007755 UP000078540 UP000000803 UP000008744 UP000036403 UP000092553 UP000076408 UP000027135 UP000078541 UP000008237 UP000005205 UP000007798 UP000001292 UP000000311 UP000001070 UP000092445 UP000245037 UP000007151 UP000075809 UP000192221 UP000235965 UP000075883 UP000075885 UP000075886 UP000009192 UP000075901 UP000008711 UP000075903 UP000001819 UP000075884 UP000075881 UP000007801 UP000002320 UP000075920 UP000095301 UP000075902 UP000008820 UP000008792 UP000007266 UP000075880 UP000076407 UP000007062 UP000095300 UP000069272 UP000002282
UP000000304 UP000078542 UP000078492 UP000218220 UP000076502 UP000053825 UP000283053 UP000268350 UP000007755 UP000078540 UP000000803 UP000008744 UP000036403 UP000092553 UP000076408 UP000027135 UP000078541 UP000008237 UP000005205 UP000007798 UP000001292 UP000000311 UP000001070 UP000092445 UP000245037 UP000007151 UP000075809 UP000192221 UP000235965 UP000075883 UP000075885 UP000075886 UP000009192 UP000075901 UP000008711 UP000075903 UP000001819 UP000075884 UP000075881 UP000007801 UP000002320 UP000075920 UP000095301 UP000075902 UP000008820 UP000008792 UP000007266 UP000075880 UP000076407 UP000007062 UP000095300 UP000069272 UP000002282
PRIDE
Pfam
PF00022 Actin
ProteinModelPortal
A0A1B0G1D5
A0A1A9W215
A0A1A9X7U1
A0A1A9UUE4
A0A1L8E5F8
A0A1L8ECP3
+ More
A0A1L8E580 A0A0L0C8P9 A0A1L8ECD3 A0A323TKK8 B4R5A1 A0A2H4H004 A0A195CW50 A0A151JNS2 A0A059XKE4 A0A1W6EVQ4 D3TQK0 E9I8G6 A0A2A4JTM4 A0A154PID2 A0A0L7QNE1 I4DLZ5 A0A3S2M069 A0A1X9H806 A0A3B0JWK6 F4WEV7 T1E2Q6 T1DEW8 A0A140F3M8 A0A195B5V2 A0A1Q3EUM5 A0A1S6Q8B9 X2JCP8 B4H0C1 Q5XUC4 A0A0J7KZ38 R4FQ90 E3UKI0 A0A0M5J8B6 A0A2R4CIH4 A0A126CN36 A0A182YH06 F2ZBU0 A0A067RMN2 D8WKR5 A0A2H4ZTI5 I4DIX7 A0A0T6B3H8 A0A2M4CSZ9 A0A1B6F0W0 A0A151K130 D0PTT4 E2BJ05 A0A158NXD4 A0A140B7Z7 B4MTB7 A0A286NL51 B4I0H7 A0A310SF14 E2AFQ5 B4JM91 A0A2M3ZF62 A0A0G2YN85 A0A1A9Z4Q1 A0A2M3ZZM2 A0A3G1T170 A0A2P8Z2J3 A0A0K8TTW7 A0A212EPG4 A0A151X9U7 A0A1W4UVV0 S5M4E2 A0A2J7PLV1 A0A182LYK9 A0A182PUA7 A0A182R1F8 B4L1G6 A0A182T7I7 A0A125RA07 B3NSY7 A0A182UTZ1 Q29H46 G3FP77 A0A182N0N1 A0A182K3Z4 A0A0P8XGW8 B0WSZ2 A0A182WQH2 A0A1I8N9U1 A0A182U3B5 Q16QR7 B4MEM3 D6WVY4 A0A1B6CYP1 A0A182J2J4 A0A182X7E8 F5HJZ8 A0A1I8PU39 A0A182FZP4 B4Q1E2
A0A1L8E580 A0A0L0C8P9 A0A1L8ECD3 A0A323TKK8 B4R5A1 A0A2H4H004 A0A195CW50 A0A151JNS2 A0A059XKE4 A0A1W6EVQ4 D3TQK0 E9I8G6 A0A2A4JTM4 A0A154PID2 A0A0L7QNE1 I4DLZ5 A0A3S2M069 A0A1X9H806 A0A3B0JWK6 F4WEV7 T1E2Q6 T1DEW8 A0A140F3M8 A0A195B5V2 A0A1Q3EUM5 A0A1S6Q8B9 X2JCP8 B4H0C1 Q5XUC4 A0A0J7KZ38 R4FQ90 E3UKI0 A0A0M5J8B6 A0A2R4CIH4 A0A126CN36 A0A182YH06 F2ZBU0 A0A067RMN2 D8WKR5 A0A2H4ZTI5 I4DIX7 A0A0T6B3H8 A0A2M4CSZ9 A0A1B6F0W0 A0A151K130 D0PTT4 E2BJ05 A0A158NXD4 A0A140B7Z7 B4MTB7 A0A286NL51 B4I0H7 A0A310SF14 E2AFQ5 B4JM91 A0A2M3ZF62 A0A0G2YN85 A0A1A9Z4Q1 A0A2M3ZZM2 A0A3G1T170 A0A2P8Z2J3 A0A0K8TTW7 A0A212EPG4 A0A151X9U7 A0A1W4UVV0 S5M4E2 A0A2J7PLV1 A0A182LYK9 A0A182PUA7 A0A182R1F8 B4L1G6 A0A182T7I7 A0A125RA07 B3NSY7 A0A182UTZ1 Q29H46 G3FP77 A0A182N0N1 A0A182K3Z4 A0A0P8XGW8 B0WSZ2 A0A182WQH2 A0A1I8N9U1 A0A182U3B5 Q16QR7 B4MEM3 D6WVY4 A0A1B6CYP1 A0A182J2J4 A0A182X7E8 F5HJZ8 A0A1I8PU39 A0A182FZP4 B4Q1E2
PDB
5WFN
E-value=0,
Score=2021
Ontologies
PATHWAY
GO
PANTHER
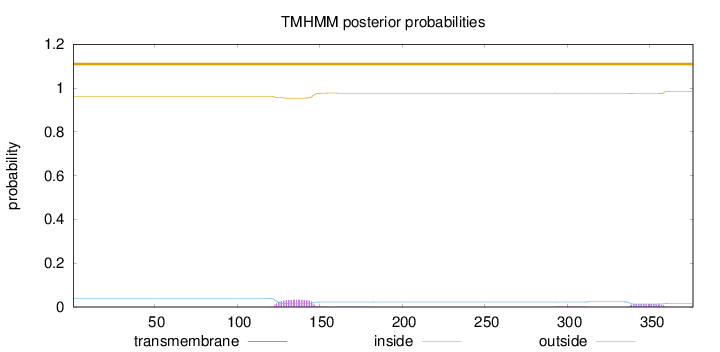
Topology
Subcellular location
Length:
376
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.00162
Exp number, first 60 AAs:
0.00114
Total prob of N-in:
0.03845
outside
1 - 376
Population Genetic Test Statistics
Pi
265.75874
Theta
194.374908
Tajima's D
1.476904
CLR
0
CSRT
0.788460576971151
Interpretation
Uncertain
