Gene
KWMTBOMO10167
Pre Gene Modal
BGIBMGA005630
Annotation
PREDICTED:_sodium/potassium-transporting_ATPase_subunit_alpha-B-like_[Papilio_xuthus]
Full name
Sodium/potassium-transporting ATPase subunit alpha
Alternative Name
Sodium pump subunit alpha
Location in the cell
PlasmaMembrane Reliability : 4.601
Sequence
CDS
ATGACTCAAATAGGTAAAATAGCAGGACTTGTGACCGGATTGAAAAAAGAAGAGACACCGATCGCTAAAGAGATAACACATTTTATCAAACTCATCTGCGGAGTTGCGTTTGTCTTTGGTATAATATTTTTTGCGATGGTGTTTTTTACGCAACACCGTTGGATATCAGCTATTCAATACATGTTAGGAATTATACTTGCCAACGTACCAGAAGGCCTGATAGTAACATTGACAGTGTGCATGACTTTGTCGGCCAACCAGCTACAAAAGAAAAACTGTTTGGCCAAAAGCTTGCACGCTGTGGAAACATTGGGCTCCACATCGTGTATTTGCTCGGACAAAACCGGTACTTTGACTGAAAACCAAATGAAGGTTTCTCATTTGTTCTGTAATTTTAAGATTTACGACAGCGACGATTTTAGTCACGTATCTCAATCTACATACAGTGCGTTGTGTTTGGCAGCTTCCCTAAACCTTAAGGCGTTTTTTGGATTTGACTACTACGACGCTCCAATTGCAAAGAGAAAGATTTATGGTGACGCTTCGGAATCTGCGATATTGCGTTACATGGAACTGCATCGTTCAGCGACGTTGACGCGCGAAGATAACCCTAAGGAGGCGGAGATTCCTTTTAGTTCAGCATACAAATACCAAGTCACGATCCATCGAATGCAAGCCTCTCAAAGCTACTATTTGACAATGAAGGGTGCTCCCGAAATTGTACTAGATTATTGCACAACTGTGCACACCGACGAAGGAGAGCAGCCGTTGACTACACAAACGATCAAAGAACTGAAAGCATACTTTGTCAAATTGGCGAATATGGGCGAACGCGTTATTGGGTATTGCGATTTGAAACTACCGACAACTGATTATCCTCTTGGATACCAATTTGACACGCATCAACTTAATTTTCCCATTGAAAATTTACACTTTCTTGGATGTATATCGATGATTGACCCCCCTAGGAAGGATATAGCGAAAAGCATATCGATGTGTAGACAAGCAGGTATCAAAGTGATCATGATAACCGGTGATCATCCTGTAACGGCATTGGCTATTTCTCGTCAGTGCGGTACCATAACACAGCCCACTGTCTACGATTATGCCTTTGAGCATCACATTGAACTGTCCGATGTTCCATCGCACATAAAAAACCAGTTCAAAGCGGCCGTGATTACAGGCGACGAGCTGCGTAAAATGTCCGTCAACGAATTGAAGTCAGCGCTAGCTAAGTACGAGGAAATAACATTCGCAAGAACCAGTCCGCAGCAGAAGCTATTCATTGTGGAGACTTATCAGTCTTTGAAGTATATCGTGGCCGTCACTGGAGACGGCGTGAACGATTCTCCTGCATTAAAGAAAGCTGATATTGGAATAGCTATGGGCATCAACGGTACAGAGGTATCAAAGCAAGCTGCAGACATGATCTTATTGGACGATAATTTTTCGTCTATTGTGTTAGGAGTGCAAGAAGGAAGACGCATTTTCGATAATTTGAAGAAAACTATAGCGTATACGCTTACCTCGAACACTCCTGAAATGCTGCCTTTCGTTATGTACGCTTGTGTCGGAATGCCTTTACCGTTAACGTTGATGTTGATTTTGGTAATCAACGTTGGAACCGACTTGTTACCAGCCATGAGTTTGGCCTACGAGGATTCCGAAGAGGACATCATGTCGATCCCTCCCAGAAAACAGACCGATCATCTCGTTAACAAAGTTCTCCTTTTCATGGCCTATTTCCAAATAGGATTAATTCAATTTTTCGCTGGAATGTATGCGTACTTTGTAGTTTTCGCCAGAGAAGGATTCTTTCCGTCTAGTTTATTGTTCGTGCGTGATGATTGGGAATCTGATACTAAAGTAATCAGAGATTCAATTGGACGGTTATATTTTTATTCGGAGCGAAAACGAATCGAAAGGAAAGCGCAGACAGCCTACTTTGTGGCTATTTGCTTAACCCAAATATCTGACGTTTTAATATGTAAGACGAGGCGGATTTCGCTCTTCAAAAAAGGCATGAAGAACCAAGTTTTGAATGCTAGTATAGTTATTGATATATTTGCAGCAGTGACTGTCACTTATTTGCCATTGTGCAACGAAATTTTCGGCACGGAGCCCATCGATTGGTATTTATTTTTCCTTGCGGTACCGTTCATGGCGCTTATGATAATTGCGGACGAGATTAGGAGGTATTTAATTAGGAATAATATTTCCAAGTGGCTTGAAAAAGAAACGTATTACTGA
Protein
MTQIGKIAGLVTGLKKEETPIAKEITHFIKLICGVAFVFGIIFFAMVFFTQHRWISAIQYMLGIILANVPEGLIVTLTVCMTLSANQLQKKNCLAKSLHAVETLGSTSCICSDKTGTLTENQMKVSHLFCNFKIYDSDDFSHVSQSTYSALCLAASLNLKAFFGFDYYDAPIAKRKIYGDASESAILRYMELHRSATLTREDNPKEAEIPFSSAYKYQVTIHRMQASQSYYLTMKGAPEIVLDYCTTVHTDEGEQPLTTQTIKELKAYFVKLANMGERVIGYCDLKLPTTDYPLGYQFDTHQLNFPIENLHFLGCISMIDPPRKDIAKSISMCRQAGIKVIMITGDHPVTALAISRQCGTITQPTVYDYAFEHHIELSDVPSHIKNQFKAAVITGDELRKMSVNELKSALAKYEEITFARTSPQQKLFIVETYQSLKYIVAVTGDGVNDSPALKKADIGIAMGINGTEVSKQAADMILLDDNFSSIVLGVQEGRRIFDNLKKTIAYTLTSNTPEMLPFVMYACVGMPLPLTLMLILVINVGTDLLPAMSLAYEDSEEDIMSIPPRKQTDHLVNKVLLFMAYFQIGLIQFFAGMYAYFVVFAREGFFPSSLLFVRDDWESDTKVIRDSIGRLYFYSERKRIERKAQTAYFVAICLTQISDVLICKTRRISLFKKGMKNQVLNASIVIDIFAAVTVTYLPLCNEIFGTEPIDWYLFFLAVPFMALMIIADEIRRYLIRNNISKWLEKETYY
Summary
Description
This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium ions, providing the energy for active transport of various nutrients.
Catalytic Activity
ATP + H2O + K(+)(out) + Na(+)(in) = ADP + H(+) + K(+)(in) + Na(+)(out) + phosphate
Subunit
The sodium/potassium-transporting ATPase is composed of a catalytic alpha subunit, an auxiliary non-catalytic beta subunit and an additional regulatory subunit.
Miscellaneous
Ouabain-sensitive electrogenic ion pump.
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IIC subfamily.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IIC subfamily.
Keywords
Alternative splicing
ATP-binding
Cell membrane
Complete proteome
Ion transport
Membrane
Nucleotide-binding
Phosphoprotein
Potassium
Potassium transport
Reference proteome
RNA editing
Sodium
Sodium transport
Translocase
Transmembrane
Transmembrane helix
Transport
Feature
chain Sodium/potassium-transporting ATPase subunit alpha
splice variant In isoform 2, isoform 3, isoform 6 and isoform 7.
splice variant In isoform 2, isoform 3, isoform 6 and isoform 7.
Uniprot
H9J7Y8
A0A2A4JSG0
A0A194QRY7
A0A0L7LF26
A0A212EST2
A0A194QB43
+ More
A0A2H1V918 A0A0P4ZDD7 A0A0P4XXA4 A0A0P4WSQ6 A0A0P6ED19 A0A0P5NDR6 A0A0P5MBV3 A0A0P5CQB2 A0A0N8A1P7 A0A0P6HW02 A0A0P5AVV6 A0A0P6FP55 A0A0P5WBW4 A0A0P5EZ23 T1KYM5 A0A0P5UV53 A0A2I9LP96 A0A0K8TRD8 A0A0R3UN08 A0A0P4ZD62 A0A0P6GTA8 T1E1A9 Q3S346 A0A0P5TZH5 Q3S347 A0A0P8YLJ0 A0A0P9C8B9 Q9GPG2 A0A2M4CUE1 B3M359 A0A131XLE6 A0A0A1WY87 K0GEX7 B3P304 A0A0A1X4H1 A0A034W2I4 A0A0D8Y0G3 A0A1I8PAR7 A0A0P5K067 A0A0K8W6J7 A0A034W3G9 B4PQ93 A0A2R5LB15 K0GEX4 A0A0K8UY58 R4VGQ9 A0A0N8P1E9 A0A034W1V5 A0A0R1E5P2 A0A2W1BK20 A0A224YZK0 K0GLY2 A0A1I8MBT1 T1PH35 K0G7C4 I7J4E4 A0A0P8XYP1 A0A0P5UVZ1 E9HAW7 A0A1I8MBT2 A9XTM7 A0A0R1E150 A0A2M4CIP8 B4JRY9 R9QXZ9 E0VNM0 P13607-4 P13607-2 A0A368H1T3 A0A1I8MBT8 A0A131YL94 A0A1I8PAR6 B4IKN2 A0A1Z5LG80 A0A0B4KGG8 A0A2A4JV07 H9ZJM5 A0A0R1E441 A0A0R1E1E2 A0A0R1E113 A0A1I8PAT4 A0A0P8YE28 A0A0N4Y050 E0VPN3 P13607 A0A1W4VZ81 A0A1I8MBT4 A0A2A4JVC3 A0A2A4JUC5 K0GM01 A0A1I8PAW2 A0A0P5CJZ8 A0A0P5C8E2 A0A0P5C931
A0A2H1V918 A0A0P4ZDD7 A0A0P4XXA4 A0A0P4WSQ6 A0A0P6ED19 A0A0P5NDR6 A0A0P5MBV3 A0A0P5CQB2 A0A0N8A1P7 A0A0P6HW02 A0A0P5AVV6 A0A0P6FP55 A0A0P5WBW4 A0A0P5EZ23 T1KYM5 A0A0P5UV53 A0A2I9LP96 A0A0K8TRD8 A0A0R3UN08 A0A0P4ZD62 A0A0P6GTA8 T1E1A9 Q3S346 A0A0P5TZH5 Q3S347 A0A0P8YLJ0 A0A0P9C8B9 Q9GPG2 A0A2M4CUE1 B3M359 A0A131XLE6 A0A0A1WY87 K0GEX7 B3P304 A0A0A1X4H1 A0A034W2I4 A0A0D8Y0G3 A0A1I8PAR7 A0A0P5K067 A0A0K8W6J7 A0A034W3G9 B4PQ93 A0A2R5LB15 K0GEX4 A0A0K8UY58 R4VGQ9 A0A0N8P1E9 A0A034W1V5 A0A0R1E5P2 A0A2W1BK20 A0A224YZK0 K0GLY2 A0A1I8MBT1 T1PH35 K0G7C4 I7J4E4 A0A0P8XYP1 A0A0P5UVZ1 E9HAW7 A0A1I8MBT2 A9XTM7 A0A0R1E150 A0A2M4CIP8 B4JRY9 R9QXZ9 E0VNM0 P13607-4 P13607-2 A0A368H1T3 A0A1I8MBT8 A0A131YL94 A0A1I8PAR6 B4IKN2 A0A1Z5LG80 A0A0B4KGG8 A0A2A4JV07 H9ZJM5 A0A0R1E441 A0A0R1E1E2 A0A0R1E113 A0A1I8PAT4 A0A0P8YE28 A0A0N4Y050 E0VPN3 P13607 A0A1W4VZ81 A0A1I8MBT4 A0A2A4JVC3 A0A2A4JUC5 K0GM01 A0A1I8PAW2 A0A0P5CJZ8 A0A0P5C8E2 A0A0P5C931
EC Number
7.2.2.13
Pubmed
19121390
26354079
26227816
22118469
29248469
26369729
+ More
17562880 17994087 18057021 11807118 28049606 25830018 23019645 25348373 26856411 17550304 28756777 28797301 25315136 21292972 20566863 2540956 9648860 10731132 12537572 12537569 12598616 9680312 2557235 17018572 26830274 28528879 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356
17562880 17994087 18057021 11807118 28049606 25830018 23019645 25348373 26856411 17550304 28756777 28797301 25315136 21292972 20566863 2540956 9648860 10731132 12537572 12537569 12598616 9680312 2557235 17018572 26830274 28528879 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01020168
NWSH01000703
PCG74706.1
KQ461155
KPJ08283.1
JTDY01001365
+ More
KOB74067.1 AGBW02012723 OWR44555.1 KQ459232 KPJ02649.1 ODYU01001126 SOQ36902.1 GDIP01218167 JAJ05235.1 GDIP01235472 JAI87929.1 GDIP01253205 JAI70196.1 GDIQ01078678 JAN16059.1 GDIQ01146812 JAL04914.1 GDIQ01162208 JAK89517.1 GDIP01168268 JAJ55134.1 GDIP01191161 JAJ32241.1 GDIQ01029055 JAN65682.1 GDIP01197974 JAJ25428.1 GDIQ01047502 JAN47235.1 GDIP01088280 JAM15435.1 GDIP01153779 JAJ69623.1 CAEY01000714 GDIP01109684 JAL94030.1 GFWZ01000215 MBW20205.1 GDAI01000902 JAI16701.1 UXSR01005651 VDD83150.1 GDIP01216015 JAJ07387.1 GDIQ01029056 JAN65681.1 GAKT01000064 JAA92998.1 DQ173925 ABA02167.1 GDIP01122237 JAL81477.1 DQ173924 ABA02166.1 CH902617 KPU79731.1 KPU79733.1 AF327439 AAG47843.1 GGFL01004280 MBW68458.1 EDV42459.1 GEFH01000672 JAP67909.1 GBXI01010944 JAD03348.1 JQ771508 AFU25677.1 CH954181 EDV48318.1 GBXI01008719 JAD05573.1 GAKP01010637 JAC48315.1 KN716280 KJH48096.1 GDIQ01191685 JAK60040.1 GDHF01005799 JAI46515.1 GAKP01010639 JAC48313.1 CM000160 EDW96202.2 GGLE01002529 MBY06655.1 JQ771506 AFU25675.1 GDHF01020773 JAI31541.1 KC691291 AGM39710.1 KPU79732.1 GAKP01010640 GAKP01010638 JAC48312.1 KRK02980.1 KZ150095 PZC73617.1 GFPF01011902 MAA23048.1 JQ771505 AFU25674.1 KA647445 AFP62074.1 JQ771507 AFU25676.1 HE962487 CCJ09645.1 KPU79736.1 GDIP01107486 JAL96228.1 GL732613 EFX71104.1 EF672699 ABV65906.1 KRK02982.1 GGFL01001054 MBW65232.1 CH916373 EDV94529.1 JX173959 AGF90965.1 AAZO01003918 DS235340 EEB14976.1 X14476 AF044974 AE014297 AY069184 AY174097 AY174098 U55767 X17471 JOJR01000021 RCN50576.1 GEDV01008478 JAP80079.1 CH480853 EDW51636.1 GFJQ02000574 JAW06396.1 AGB96170.1 NWSH01000562 PCG75639.1 BT133381 AFH36366.1 KRK02979.1 KRK02985.1 KRK02987.1 KPU79735.1 UYSL01020061 VDL72459.1 AAZO01004204 DS235366 EEB15339.1 PCG75638.1 PCG75637.1 JQ771525 AFU25694.1 GDIP01174410 JAJ48992.1 GDIP01174412 JAJ48990.1 GDIP01174411 JAJ48991.1
KOB74067.1 AGBW02012723 OWR44555.1 KQ459232 KPJ02649.1 ODYU01001126 SOQ36902.1 GDIP01218167 JAJ05235.1 GDIP01235472 JAI87929.1 GDIP01253205 JAI70196.1 GDIQ01078678 JAN16059.1 GDIQ01146812 JAL04914.1 GDIQ01162208 JAK89517.1 GDIP01168268 JAJ55134.1 GDIP01191161 JAJ32241.1 GDIQ01029055 JAN65682.1 GDIP01197974 JAJ25428.1 GDIQ01047502 JAN47235.1 GDIP01088280 JAM15435.1 GDIP01153779 JAJ69623.1 CAEY01000714 GDIP01109684 JAL94030.1 GFWZ01000215 MBW20205.1 GDAI01000902 JAI16701.1 UXSR01005651 VDD83150.1 GDIP01216015 JAJ07387.1 GDIQ01029056 JAN65681.1 GAKT01000064 JAA92998.1 DQ173925 ABA02167.1 GDIP01122237 JAL81477.1 DQ173924 ABA02166.1 CH902617 KPU79731.1 KPU79733.1 AF327439 AAG47843.1 GGFL01004280 MBW68458.1 EDV42459.1 GEFH01000672 JAP67909.1 GBXI01010944 JAD03348.1 JQ771508 AFU25677.1 CH954181 EDV48318.1 GBXI01008719 JAD05573.1 GAKP01010637 JAC48315.1 KN716280 KJH48096.1 GDIQ01191685 JAK60040.1 GDHF01005799 JAI46515.1 GAKP01010639 JAC48313.1 CM000160 EDW96202.2 GGLE01002529 MBY06655.1 JQ771506 AFU25675.1 GDHF01020773 JAI31541.1 KC691291 AGM39710.1 KPU79732.1 GAKP01010640 GAKP01010638 JAC48312.1 KRK02980.1 KZ150095 PZC73617.1 GFPF01011902 MAA23048.1 JQ771505 AFU25674.1 KA647445 AFP62074.1 JQ771507 AFU25676.1 HE962487 CCJ09645.1 KPU79736.1 GDIP01107486 JAL96228.1 GL732613 EFX71104.1 EF672699 ABV65906.1 KRK02982.1 GGFL01001054 MBW65232.1 CH916373 EDV94529.1 JX173959 AGF90965.1 AAZO01003918 DS235340 EEB14976.1 X14476 AF044974 AE014297 AY069184 AY174097 AY174098 U55767 X17471 JOJR01000021 RCN50576.1 GEDV01008478 JAP80079.1 CH480853 EDW51636.1 GFJQ02000574 JAW06396.1 AGB96170.1 NWSH01000562 PCG75639.1 BT133381 AFH36366.1 KRK02979.1 KRK02985.1 KRK02987.1 KPU79735.1 UYSL01020061 VDL72459.1 AAZO01004204 DS235366 EEB15339.1 PCG75638.1 PCG75637.1 JQ771525 AFU25694.1 GDIP01174410 JAJ48992.1 GDIP01174412 JAJ48990.1 GDIP01174411 JAJ48991.1
Proteomes
Interpro
IPR004014
ATPase_P-typ_cation-transptr_N
+ More
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR006068 ATPase_P-typ_cation-transptr_C
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR036412 HAD-like_sf
IPR023214 HAD_sf
IPR005775 P-type_ATPase_IIC
IPR018303 ATPase_P-typ_P_site
IPR031311 CHIT_BIND_RR_consensus
IPR000618 Insect_cuticle
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR006068 ATPase_P-typ_cation-transptr_C
IPR023298 ATPase_P-typ_TM_dom_sf
IPR001757 P_typ_ATPase
IPR023299 ATPase_P-typ_cyto_dom_N
IPR036412 HAD-like_sf
IPR023214 HAD_sf
IPR005775 P-type_ATPase_IIC
IPR018303 ATPase_P-typ_P_site
IPR031311 CHIT_BIND_RR_consensus
IPR000618 Insect_cuticle
Gene 3D
ProteinModelPortal
H9J7Y8
A0A2A4JSG0
A0A194QRY7
A0A0L7LF26
A0A212EST2
A0A194QB43
+ More
A0A2H1V918 A0A0P4ZDD7 A0A0P4XXA4 A0A0P4WSQ6 A0A0P6ED19 A0A0P5NDR6 A0A0P5MBV3 A0A0P5CQB2 A0A0N8A1P7 A0A0P6HW02 A0A0P5AVV6 A0A0P6FP55 A0A0P5WBW4 A0A0P5EZ23 T1KYM5 A0A0P5UV53 A0A2I9LP96 A0A0K8TRD8 A0A0R3UN08 A0A0P4ZD62 A0A0P6GTA8 T1E1A9 Q3S346 A0A0P5TZH5 Q3S347 A0A0P8YLJ0 A0A0P9C8B9 Q9GPG2 A0A2M4CUE1 B3M359 A0A131XLE6 A0A0A1WY87 K0GEX7 B3P304 A0A0A1X4H1 A0A034W2I4 A0A0D8Y0G3 A0A1I8PAR7 A0A0P5K067 A0A0K8W6J7 A0A034W3G9 B4PQ93 A0A2R5LB15 K0GEX4 A0A0K8UY58 R4VGQ9 A0A0N8P1E9 A0A034W1V5 A0A0R1E5P2 A0A2W1BK20 A0A224YZK0 K0GLY2 A0A1I8MBT1 T1PH35 K0G7C4 I7J4E4 A0A0P8XYP1 A0A0P5UVZ1 E9HAW7 A0A1I8MBT2 A9XTM7 A0A0R1E150 A0A2M4CIP8 B4JRY9 R9QXZ9 E0VNM0 P13607-4 P13607-2 A0A368H1T3 A0A1I8MBT8 A0A131YL94 A0A1I8PAR6 B4IKN2 A0A1Z5LG80 A0A0B4KGG8 A0A2A4JV07 H9ZJM5 A0A0R1E441 A0A0R1E1E2 A0A0R1E113 A0A1I8PAT4 A0A0P8YE28 A0A0N4Y050 E0VPN3 P13607 A0A1W4VZ81 A0A1I8MBT4 A0A2A4JVC3 A0A2A4JUC5 K0GM01 A0A1I8PAW2 A0A0P5CJZ8 A0A0P5C8E2 A0A0P5C931
A0A2H1V918 A0A0P4ZDD7 A0A0P4XXA4 A0A0P4WSQ6 A0A0P6ED19 A0A0P5NDR6 A0A0P5MBV3 A0A0P5CQB2 A0A0N8A1P7 A0A0P6HW02 A0A0P5AVV6 A0A0P6FP55 A0A0P5WBW4 A0A0P5EZ23 T1KYM5 A0A0P5UV53 A0A2I9LP96 A0A0K8TRD8 A0A0R3UN08 A0A0P4ZD62 A0A0P6GTA8 T1E1A9 Q3S346 A0A0P5TZH5 Q3S347 A0A0P8YLJ0 A0A0P9C8B9 Q9GPG2 A0A2M4CUE1 B3M359 A0A131XLE6 A0A0A1WY87 K0GEX7 B3P304 A0A0A1X4H1 A0A034W2I4 A0A0D8Y0G3 A0A1I8PAR7 A0A0P5K067 A0A0K8W6J7 A0A034W3G9 B4PQ93 A0A2R5LB15 K0GEX4 A0A0K8UY58 R4VGQ9 A0A0N8P1E9 A0A034W1V5 A0A0R1E5P2 A0A2W1BK20 A0A224YZK0 K0GLY2 A0A1I8MBT1 T1PH35 K0G7C4 I7J4E4 A0A0P8XYP1 A0A0P5UVZ1 E9HAW7 A0A1I8MBT2 A9XTM7 A0A0R1E150 A0A2M4CIP8 B4JRY9 R9QXZ9 E0VNM0 P13607-4 P13607-2 A0A368H1T3 A0A1I8MBT8 A0A131YL94 A0A1I8PAR6 B4IKN2 A0A1Z5LG80 A0A0B4KGG8 A0A2A4JV07 H9ZJM5 A0A0R1E441 A0A0R1E1E2 A0A0R1E113 A0A1I8PAT4 A0A0P8YE28 A0A0N4Y050 E0VPN3 P13607 A0A1W4VZ81 A0A1I8MBT4 A0A2A4JVC3 A0A2A4JUC5 K0GM01 A0A1I8PAW2 A0A0P5CJZ8 A0A0P5C8E2 A0A0P5C931
PDB
3WGV
E-value=0,
Score=1850
Ontologies
KEGG
GO
GO:0005524
GO:0016021
GO:0008556
GO:0046872
GO:0042302
GO:0009612
GO:0050905
GO:0019991
GO:0008340
GO:0035158
GO:0009266
GO:0007605
GO:0005634
GO:0051124
GO:0001894
GO:0007268
GO:0016323
GO:0060439
GO:0005391
GO:0005918
GO:0007630
GO:0006883
GO:0030007
GO:0005623
GO:0015991
GO:0036376
GO:1990573
GO:0005890
GO:0008344
GO:0006812
GO:0005737
GO:0007626
GO:0035159
GO:0005886
GO:0000166
GO:0008270
GO:0042773
GO:0016651
GO:0006402
GO:0003824
Topology
Subcellular location
Membrane
Cell membrane
Cell membrane
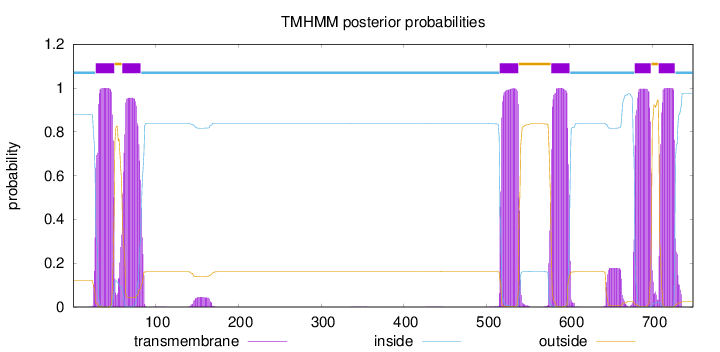
Length:
749
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
134.31717
Exp number, first 60 AAs:
23.51722
Total prob of N-in:
0.87923
POSSIBLE N-term signal
sequence
inside
1 - 27
TMhelix
28 - 50
outside
51 - 59
TMhelix
60 - 82
inside
83 - 515
TMhelix
516 - 538
outside
539 - 577
TMhelix
578 - 600
inside
601 - 678
TMhelix
679 - 698
outside
699 - 707
TMhelix
708 - 727
inside
728 - 749
Population Genetic Test Statistics
Pi
229.310996
Theta
192.441454
Tajima's D
0.456993
CLR
0.056902
CSRT
0.500174991250438
Interpretation
Uncertain
