Gene
KWMTBOMO10164
Pre Gene Modal
BGIBMGA005572
Annotation
PREDICTED:_intraflagellar_transport_protein_56?_partial_[Papilio_polytes]
Full name
Intraflagellar transport protein 56
Alternative Name
Tetratricopeptide repeat protein 26
Location in the cell
Cytoplasmic Reliability : 1.447 Nuclear Reliability : 1.601
Sequence
CDS
ATGGATGAAAGAATAGCTGATAATATAGAAATGGATATTGCTGTGTGCTATTTCTATTTAGGGATGTACAAGGAATCGCATGAAGCCGCCGAAAAAGCTCCAACATGCACCTTGAAGAACCGACTACAGTTCCACTTGGCTCACAAGCTCAACGACGAGGAAGGTCTGATGCGTACTCACGCTGCCCTTCGAGATATACCGGAAGACCAGCTCAGCTTGGCCTCAGTTCACTACCTGAGGGCTCATTACCAGGAAGCTATTGATGTTTACAAGAAATTGTTGCTCGAAAGACGGTAG
Protein
MDERIADNIEMDIAVCYFYLGMYKESHEAAEKAPTCTLKNRLQFHLAHKLNDEEGLMRTHAALRDIPEDQLSLASVHYLRAHYQEAIDVYKKLLLERR
Summary
Description
Component of the intraflagellar transport (IFT) complex B required for transport of proteins in the motile cilium. Required for transport of specific ciliary cargo proteins related to motility, while it is neither required for IFT complex B assembly or motion nor for cilium assembly. Plays a key role in maintaining the integrity of the IFT complex B and the proper ciliary localization of the IFT complex B components. Essential for maintaining proper microtubule organization within the ciliary axoneme.
Subunit
Component of the IFT complex B.
Similarity
Belongs to the IFT56 family.
Keywords
Cell projection
Cilium
Complete proteome
Protein transport
Reference proteome
Repeat
TPR repeat
Transport
Feature
chain Intraflagellar transport protein 56
Uniprot
H9J7T0
A0A2A4JSW2
A0A212F6I1
A0A194QAT4
A0A194QTF4
A0A2H1W9I1
+ More
A0A2H1V7R7 D0ABA5 A7SZB0 A0A1S8W947 A0A1S8W958 A0A1Y2C711 U4U800 N6UGX7 A0A177WHF2 F4PE37 A0A1L8GND8 A0A0T6B3F9 A0A0L0HEW4 A0A1S3HKT6 A0A1S3KIA4 A0A0L0HE99 A0A1Y1KJ10 A0A1B0DNX6 A0A091K2U2 A0A1Y1VJ22 A0A2B4RLZ5 A0A3M6TES6 A0A1Y1WW27 A0A0L0T2R8 C3Y9Y7 A0A3Q1NHN6 D6WVY7 A0A2Y9TAM0 A0A3Q1MHS6 A0A287AI77 A0A2Y9TAL4 F1MF33 K9KDE1 A0A210QW50 F6SI03 A0A088AR85 L8IBA0 A0A2Y9T774 A0A091KDX0 A0A369RSR6 B3S987 A0A1V4J9V8 A0A2U4CFQ4 G1TEP6 A0A091W1S0 A0A384CLA1 A0A3Q7XLY9 A0A138ZZD7 A0A093SXL7 A0A2U4CFA2 A0A151HXU3 V9KR02 A0A2U4CF51 A0A1V4JAA7 G1NHH2 W5N287 G3TCC1 A0A0F8AT85 A0A2U4CF47 A4III8 A0A341B9E4 F7BZC2 A0A2I0MJE2 A0A1U7TV60 A0A2A3E9E2 A0A1S3WE55 R7VWU5 A0A3P4PBT3 A0A093BHZ9 A0A341B7X5 A0A2U4CEG8 A0A093QN25 A0A060VXV7 A0A2U3XWU1 A0A0M8ZUM4 A0A091HLF9 A0A1S3S3W4 A0A2I0USD4 A0A3Q7QV35 H2ZMQ2 A0A093FYU8 A0A1W4WFA0 A0A1S3S3X2 A0A337SHK5 A0A2U3WXA4 A0A2Y9NL40 A0A1S3L7D5 A0A094MY89 A0A337SKE0 U6D590 A0A3Q2H7I7 A0A091SYS2 A0A060XL21 A0A151WI44
A0A2H1V7R7 D0ABA5 A7SZB0 A0A1S8W947 A0A1S8W958 A0A1Y2C711 U4U800 N6UGX7 A0A177WHF2 F4PE37 A0A1L8GND8 A0A0T6B3F9 A0A0L0HEW4 A0A1S3HKT6 A0A1S3KIA4 A0A0L0HE99 A0A1Y1KJ10 A0A1B0DNX6 A0A091K2U2 A0A1Y1VJ22 A0A2B4RLZ5 A0A3M6TES6 A0A1Y1WW27 A0A0L0T2R8 C3Y9Y7 A0A3Q1NHN6 D6WVY7 A0A2Y9TAM0 A0A3Q1MHS6 A0A287AI77 A0A2Y9TAL4 F1MF33 K9KDE1 A0A210QW50 F6SI03 A0A088AR85 L8IBA0 A0A2Y9T774 A0A091KDX0 A0A369RSR6 B3S987 A0A1V4J9V8 A0A2U4CFQ4 G1TEP6 A0A091W1S0 A0A384CLA1 A0A3Q7XLY9 A0A138ZZD7 A0A093SXL7 A0A2U4CFA2 A0A151HXU3 V9KR02 A0A2U4CF51 A0A1V4JAA7 G1NHH2 W5N287 G3TCC1 A0A0F8AT85 A0A2U4CF47 A4III8 A0A341B9E4 F7BZC2 A0A2I0MJE2 A0A1U7TV60 A0A2A3E9E2 A0A1S3WE55 R7VWU5 A0A3P4PBT3 A0A093BHZ9 A0A341B7X5 A0A2U4CEG8 A0A093QN25 A0A060VXV7 A0A2U3XWU1 A0A0M8ZUM4 A0A091HLF9 A0A1S3S3W4 A0A2I0USD4 A0A3Q7QV35 H2ZMQ2 A0A093FYU8 A0A1W4WFA0 A0A1S3S3X2 A0A337SHK5 A0A2U3WXA4 A0A2Y9NL40 A0A1S3L7D5 A0A094MY89 A0A337SKE0 U6D590 A0A3Q2H7I7 A0A091SYS2 A0A060XL21 A0A151WI44
Pubmed
EMBL
BABH01020169
BABH01020170
NWSH01000703
PCG74704.1
AGBW02010007
OWR49346.1
+ More
KQ459232 KPJ02648.1 KQ461155 KPJ08285.1 ODYU01007198 SOQ49765.1 ODYU01001126 SOQ36903.1 FP102341 CBH09300.1 DS469957 EDO30962.1 LYON01000022 OON10992.1 OON10991.1 MCGO01000027 ORY42813.1 KB632064 ERL88453.1 APGK01035650 APGK01035651 KB740928 ENN77902.1 DS022302 OAJ39206.1 GL882896 EGF76485.1 CM004472 OCT85347.1 LJIG01016001 KRT81913.1 KQ257458 KNC99308.1 KNC99309.1 GEZM01082174 JAV61413.1 AJVK01007813 AJVK01007814 KK544172 KFP31792.1 MCFH01000008 ORX56092.1 LSMT01000446 PFX17839.1 RCHS01003767 RMX39882.1 MCFG01000235 ORX77761.1 GG745358 KNE68899.1 GG666493 EEN62896.1 KQ971359 EFA08635.1 AEMK02000111 JL623455 AEP99724.1 NEDP02001549 OWF52990.1 JH881572 ELR53498.1 KK741016 KFP38774.1 NOWV01000219 RDD37735.1 DS985258 EDV20604.1 LSYS01008398 OPJ69018.1 KL411538 KFR08960.1 KQ965850 KXS09778.1 KL672898 KFW87269.1 KQ976748 KYM75317.1 JW868250 AFP00768.1 OPJ69019.1 AHAT01036115 AHAT01036116 AHAT01036117 AHAT01036118 AHAT01036119 AHAT01036120 AHAT01036121 AHAT01036122 AHAT01036123 AHAT01036124 KQ042803 KKF10466.1 BC136035 AAMC01036707 AAMC01036708 AKCR02000009 PKK29788.1 KZ288313 PBC28345.1 KB375573 EMC84808.1 PKK29789.1 CYRY02031824 VCX07593.1 KN125811 KFU84543.1 KL426006 KFW90303.1 FR904335 CDQ59848.1 KQ435837 KOX71475.1 KL217504 KFO96044.1 KZ505643 PKU48949.1 KK637963 KFV59571.1 AANG04000096 KL260558 KFZ59446.1 HAAF01006336 CCP78160.1 KK489876 KFQ63160.1 FR905579 CDQ80258.1 KQ983097 KYQ47498.1
KQ459232 KPJ02648.1 KQ461155 KPJ08285.1 ODYU01007198 SOQ49765.1 ODYU01001126 SOQ36903.1 FP102341 CBH09300.1 DS469957 EDO30962.1 LYON01000022 OON10992.1 OON10991.1 MCGO01000027 ORY42813.1 KB632064 ERL88453.1 APGK01035650 APGK01035651 KB740928 ENN77902.1 DS022302 OAJ39206.1 GL882896 EGF76485.1 CM004472 OCT85347.1 LJIG01016001 KRT81913.1 KQ257458 KNC99308.1 KNC99309.1 GEZM01082174 JAV61413.1 AJVK01007813 AJVK01007814 KK544172 KFP31792.1 MCFH01000008 ORX56092.1 LSMT01000446 PFX17839.1 RCHS01003767 RMX39882.1 MCFG01000235 ORX77761.1 GG745358 KNE68899.1 GG666493 EEN62896.1 KQ971359 EFA08635.1 AEMK02000111 JL623455 AEP99724.1 NEDP02001549 OWF52990.1 JH881572 ELR53498.1 KK741016 KFP38774.1 NOWV01000219 RDD37735.1 DS985258 EDV20604.1 LSYS01008398 OPJ69018.1 KL411538 KFR08960.1 KQ965850 KXS09778.1 KL672898 KFW87269.1 KQ976748 KYM75317.1 JW868250 AFP00768.1 OPJ69019.1 AHAT01036115 AHAT01036116 AHAT01036117 AHAT01036118 AHAT01036119 AHAT01036120 AHAT01036121 AHAT01036122 AHAT01036123 AHAT01036124 KQ042803 KKF10466.1 BC136035 AAMC01036707 AAMC01036708 AKCR02000009 PKK29788.1 KZ288313 PBC28345.1 KB375573 EMC84808.1 PKK29789.1 CYRY02031824 VCX07593.1 KN125811 KFU84543.1 KL426006 KFW90303.1 FR904335 CDQ59848.1 KQ435837 KOX71475.1 KL217504 KFO96044.1 KZ505643 PKU48949.1 KK637963 KFV59571.1 AANG04000096 KL260558 KFZ59446.1 HAAF01006336 CCP78160.1 KK489876 KFQ63160.1 FR905579 CDQ80258.1 KQ983097 KYQ47498.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000001593
+ More
UP000189906 UP000193642 UP000030742 UP000019118 UP000077115 UP000007241 UP000186698 UP000053201 UP000085678 UP000092462 UP000193719 UP000225706 UP000275408 UP000193944 UP000054350 UP000001554 UP000009136 UP000007266 UP000008227 UP000242188 UP000002281 UP000005203 UP000248484 UP000253843 UP000009022 UP000190648 UP000245320 UP000001811 UP000053283 UP000261680 UP000286642 UP000070544 UP000053258 UP000078540 UP000001645 UP000018468 UP000007646 UP000008143 UP000252040 UP000053872 UP000189704 UP000242457 UP000079721 UP000193380 UP000245341 UP000053105 UP000054308 UP000087266 UP000286640 UP000007875 UP000192223 UP000011712 UP000245340 UP000248483 UP000075809
UP000189906 UP000193642 UP000030742 UP000019118 UP000077115 UP000007241 UP000186698 UP000053201 UP000085678 UP000092462 UP000193719 UP000225706 UP000275408 UP000193944 UP000054350 UP000001554 UP000009136 UP000007266 UP000008227 UP000242188 UP000002281 UP000005203 UP000248484 UP000253843 UP000009022 UP000190648 UP000245320 UP000001811 UP000053283 UP000261680 UP000286642 UP000070544 UP000053258 UP000078540 UP000001645 UP000018468 UP000007646 UP000008143 UP000252040 UP000053872 UP000189704 UP000242457 UP000079721 UP000193380 UP000245341 UP000053105 UP000054308 UP000087266 UP000286640 UP000007875 UP000192223 UP000011712 UP000245340 UP000248483 UP000075809
PRIDE
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9J7T0
A0A2A4JSW2
A0A212F6I1
A0A194QAT4
A0A194QTF4
A0A2H1W9I1
+ More
A0A2H1V7R7 D0ABA5 A7SZB0 A0A1S8W947 A0A1S8W958 A0A1Y2C711 U4U800 N6UGX7 A0A177WHF2 F4PE37 A0A1L8GND8 A0A0T6B3F9 A0A0L0HEW4 A0A1S3HKT6 A0A1S3KIA4 A0A0L0HE99 A0A1Y1KJ10 A0A1B0DNX6 A0A091K2U2 A0A1Y1VJ22 A0A2B4RLZ5 A0A3M6TES6 A0A1Y1WW27 A0A0L0T2R8 C3Y9Y7 A0A3Q1NHN6 D6WVY7 A0A2Y9TAM0 A0A3Q1MHS6 A0A287AI77 A0A2Y9TAL4 F1MF33 K9KDE1 A0A210QW50 F6SI03 A0A088AR85 L8IBA0 A0A2Y9T774 A0A091KDX0 A0A369RSR6 B3S987 A0A1V4J9V8 A0A2U4CFQ4 G1TEP6 A0A091W1S0 A0A384CLA1 A0A3Q7XLY9 A0A138ZZD7 A0A093SXL7 A0A2U4CFA2 A0A151HXU3 V9KR02 A0A2U4CF51 A0A1V4JAA7 G1NHH2 W5N287 G3TCC1 A0A0F8AT85 A0A2U4CF47 A4III8 A0A341B9E4 F7BZC2 A0A2I0MJE2 A0A1U7TV60 A0A2A3E9E2 A0A1S3WE55 R7VWU5 A0A3P4PBT3 A0A093BHZ9 A0A341B7X5 A0A2U4CEG8 A0A093QN25 A0A060VXV7 A0A2U3XWU1 A0A0M8ZUM4 A0A091HLF9 A0A1S3S3W4 A0A2I0USD4 A0A3Q7QV35 H2ZMQ2 A0A093FYU8 A0A1W4WFA0 A0A1S3S3X2 A0A337SHK5 A0A2U3WXA4 A0A2Y9NL40 A0A1S3L7D5 A0A094MY89 A0A337SKE0 U6D590 A0A3Q2H7I7 A0A091SYS2 A0A060XL21 A0A151WI44
A0A2H1V7R7 D0ABA5 A7SZB0 A0A1S8W947 A0A1S8W958 A0A1Y2C711 U4U800 N6UGX7 A0A177WHF2 F4PE37 A0A1L8GND8 A0A0T6B3F9 A0A0L0HEW4 A0A1S3HKT6 A0A1S3KIA4 A0A0L0HE99 A0A1Y1KJ10 A0A1B0DNX6 A0A091K2U2 A0A1Y1VJ22 A0A2B4RLZ5 A0A3M6TES6 A0A1Y1WW27 A0A0L0T2R8 C3Y9Y7 A0A3Q1NHN6 D6WVY7 A0A2Y9TAM0 A0A3Q1MHS6 A0A287AI77 A0A2Y9TAL4 F1MF33 K9KDE1 A0A210QW50 F6SI03 A0A088AR85 L8IBA0 A0A2Y9T774 A0A091KDX0 A0A369RSR6 B3S987 A0A1V4J9V8 A0A2U4CFQ4 G1TEP6 A0A091W1S0 A0A384CLA1 A0A3Q7XLY9 A0A138ZZD7 A0A093SXL7 A0A2U4CFA2 A0A151HXU3 V9KR02 A0A2U4CF51 A0A1V4JAA7 G1NHH2 W5N287 G3TCC1 A0A0F8AT85 A0A2U4CF47 A4III8 A0A341B9E4 F7BZC2 A0A2I0MJE2 A0A1U7TV60 A0A2A3E9E2 A0A1S3WE55 R7VWU5 A0A3P4PBT3 A0A093BHZ9 A0A341B7X5 A0A2U4CEG8 A0A093QN25 A0A060VXV7 A0A2U3XWU1 A0A0M8ZUM4 A0A091HLF9 A0A1S3S3W4 A0A2I0USD4 A0A3Q7QV35 H2ZMQ2 A0A093FYU8 A0A1W4WFA0 A0A1S3S3X2 A0A337SHK5 A0A2U3WXA4 A0A2Y9NL40 A0A1S3L7D5 A0A094MY89 A0A337SKE0 U6D590 A0A3Q2H7I7 A0A091SYS2 A0A060XL21 A0A151WI44
Ontologies
KEGG
GO
GO:0030992
GO:0036064
GO:0097546
GO:0035735
GO:0120170
GO:0035720
GO:0031514
GO:1905198
GO:0061512
GO:0035082
GO:0007224
GO:0005813
GO:0003727
GO:0032197
GO:1990904
GO:0005634
GO:0042073
GO:0005929
GO:0035845
GO:0008594
GO:0039023
GO:0005515
GO:0003676
GO:0008270
GO:0042773
GO:0016651
GO:0006402
GO:0046872
GO:0003824
PANTHER
Topology
Subcellular location
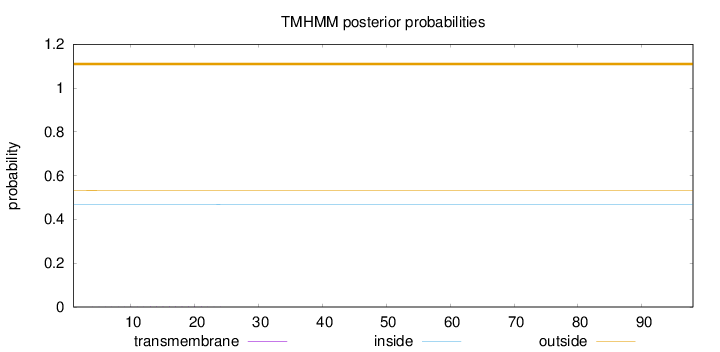
Length:
98
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02419
Exp number, first 60 AAs:
0.02419
Total prob of N-in:
0.46807
outside
1 - 98
Population Genetic Test Statistics
Pi
13.60528
Theta
15.154764
Tajima's D
-0.600485
CLR
3.196425
CSRT
0.215239238038098
Interpretation
Uncertain
