Gene
KWMTBOMO10159 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005570
Annotation
PREDICTED:_lethal(2)_giant_larvae_protein_isoform_X2_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 2.734
Sequence
CDS
ATGGCAGGCAGAATGTTGAAGTTTATACGCGGCAAGGGCCAGCAGCCCTCTGCTGAGAGGCAGAAGTTGCAGAATGAACTATTTGCATTCCGAAAGACAGTCCAGCATGGCTTCCCGCACAGAGCGTCAGCACTCGCGTGGGATCCGCTCCTCAGACTCGCGGCGCTTGGTACCGCCACCGGAGCGCTCAAGGTCTACGGCAGACCAGGCGTCGAACTCTACGGCCAACACACCAATTACGAATCGATAGTCACACAGATACATTTCATCACAGGGACAGGCCGTTTGATATCATTATGCGACGATAACAGCCTTCACTTGTGGGAGATCAACGAGAAATCTCTAGTGGAATTAAAGTCGCATGTATTCGAAGGTAAAAATAAAAAGATATCGGCCATATGCGTAGAATCGTCAGCGAAGAACGCTTTACTGGGAACCGAAGGCGGCAACATTTACTCATTGGATTTGAGCGCTTTCACGATCTCGGAAGATGTCATATACCAGGATGTCGTTATGCAGAACTGCCCAGAGGATTACAAAGTTAACCCAGGCGCGGTGGAGTCTGTATACGAACACCCTAAGACTCCTACACGTATTCTGATCGGATACAACCGAGGACTGGCCGTGCTTTGGGACCGGGAGGCTGCGGCTCCCACACATACATTTGTTTCGAATCAACAGCTTGAAAGCTTATGCTGGAATGACGATGGCGAGCACTTCACATCATCGCACAACGACGGTTCGTACGTGACGTGGGAGGTGGCCGGCGCCTCCAGCGACCGACCCCTGAAGGAGCCCGTCACGCCCTACGGACCCTACCCTTGTAAGGCGATCACCAAGATCCTGAACAGGACTAGCGTTGATGGAGAGGACATCACTATATTTGCCGGTGGCATGCCCAGAGCTTCGTACTCCGACAAGTACACGGTTACAGTGCAGCAGGGCGAGAAACATGTGGCCTTCGACTTCACGAGCCGGGTTATAGATTTGTTCACAACGACCCCCGTGCCCCCGGACGGCGCGCCGCTGCAGGAGGAGCAGCCCGACACCCCGGCCCTCGTACAGGTGCAGGCGGTCAACCAGGTCGCCGCCGCTTTGGTGGTCCTGGCGGAAGAGGAGCTGGTGGTGATAGACCTGTGCGACGCGCGCTGGCGGCCGCTGCGGCTGCCGTACCTGGTGTCGGTGCACGCGTCCGCCGTCACCACGGCGCAGCTGGCCGACGGCGTGGCGCCCGCAGTCTACGACAACATCGTCGCCGCGGGTCAACAACAAACGGAGAATCAATACTCTGAAAGTCCGTGGCCGATTTCCGGTGGGGTGGTGGAGACGGCGGAGCTGACCGACAGACAGGTGCTGCTCACGGGGCACGAGGACGGCTCCGTGCGGTTCTGGGACGTGACCGGCGTCGCCATGACCCCCCTGTACAAGTACACCACCGCGCAGCTGTTCAGTGGTGAAGAACTTGGTGAAAACAACGACAGTCAGACCACAGATGAAGAGGAGTGGCCTCCCTTCAGGCGAGTCGGCACCTTCGACCCGTACAGCGACGACCCCCGGCTCGCGGTCAAAAAGGTTATATTGTGCCCGTTATCTGGAATGCTCACCATCGGCGGAGCCGCCGGCCATATTGTCATAGCCAGTCTCAAGACCTCTCCGAGCACCACCGAAGTCAAGTCGGTGTCAGTGAATATAGTGTCAGACAGAGACGGCTTCGTGTGGAAGGGACACGGGCAGCTCACGTTGAGGGCCGGCACGCACACCTTCCCGCAGGGATATCAGGCGACCTCGGTGGTGCAGCTGCAGCCCCCGGCGGCCGTGACGTCACTGAGCGCGCAGTGGGAGTGGGGCGTGGTGTGCGCCGGCACCGCGCACGGGCTGGCGCTGGTGGACGCGCTGGGCCCGGCACCCGCGCCGCTGCTGCATAAGTGCACGCTCAACCCGCACGATCACGGATCCGGTGACACGCCAATATCGAGAAGAAAGTCCTTCAAGAAATCCTTGAGAGAGTCATTCAGGAGGCTTCGTAAAGGACGTTCACAACGGCGACAGACGACATCGAATCCCACCTCGCCCACACAACCGATTCCGAAGAAGCCAATTGACAAAGCCGCCTCGGAAACGGACGCGGAAATCAAACCGATCGAGCGCGCGGTGGAGGCCCGCTCCACGGACGACGCCTACGGCTCGATGGTGCGCTGCCTGTACTTCGCGAGGACCTTCCTCATCAATACCCAGCAATCGACGCCGACGCTATGGGCCGGCACGAACAACGGCACTGTGTACGCCTTCACGCTGCACGTGCCCAACACTAACAAACGGAAAGAGGAACCAGTAACGTGTCAGCTAGCGAAAGAAATACAGTTGAAGCACAGGGCCCCGGTCATAGGGATCACAGTACTCGACGGCGCCTCCGTACCTCTGCCCGATCCCCTAGAGGTGGAACGAGGTGTGTCCCCGCTACCGGAGCCGGGTACACATCGTGTAGTCATCACGTCGGAGGAACAGTTCAAGATATTCACTCTGCCTTCGTTGAAACCGCACAACAAGTACAAGCTGACCGCGCACGAGGGCGCTCGGGTTCGGCGCACGGCGTGGTCGCAGTTCTCGTGCGGCGCGCACCGGGAGTGGTGCTTGCTGTGCCTCACCAACCTCGGCGACTGCCTCGTGCTCAGCCCGGAGCTGCGCCGCCAGCTCAACGCCGCCGCCGTCAGGAAGGAGGACATCAATGGGATTTCGAGTTTATGTTTCTCGAAACGAGGAGAAGCTCTCTACCTGCACTCGTCTTCCGAATTACAACGGATTACTTTATCTGCTACGAAGGTGACGATCGCGGAGTGCCACGTGCTGTTGGCGCCCTGGGCGGCCGCGCTGCGGGGGCCCGCGCCGGAGCCGCCGCTCACCAACGGGGAGCATAAGGTAGACTGTGTACTCATAGTGGGATTGACGACACAGGAAGAGGTGCCGGAGACAGCTCATGACGTCACGGCGGCCTCGGCGGACATCACCGTGGACAGCGTGCGCGACCACACCAGCGCGGAGCACCCGCCCGAACTCAACATCAACTTGCAGAACACGCAAGTGAATACGAGTTCGATGGTAATAAAGACAACGACGCGCACAACGGTGAACGAGACCAACGCGGACGGCACCACCACCACCACCACCACGACCAGCACTAACAACGCCAACGAGAACATCCTAGAGCACAGTCGCGAGGAGGGAGTCATCACCCGCATCGAGACGGGAACCGTGACGGTGCCGGCCGGCACAGATCCGAAGCTGATCCTCGAGATGTTTGATCGACAACGGTCCCCCCTGGCGGTTGTTAAAAATCCAACCATTACAACAAAACTTTGTGCAAAATGCTACAGAAAAATATTATCATTTCATAAATTTAAATCGCTGGCCATAGAAAATGATATTTATTTAGAAAGTCAGTGTACATCTGCCAAGCACCAGATTCCACTCAATAACAATGAGAATGAGATTGGGCCCACACAACAGTTTAGTATACAAATGGTTGACTGTCTCTCTCCTGAAATGAACAACAAAGATATAATATTCAGAGAACAAGTCAAGACAGAAATGGTATTGGCTCATGATGAAGAAAATGACAGTATCGAGTTAGGAAAATCAGAGCCTCCAGAGTTAGAGTCTGATGATGAATATTTGAGTGTAATTAAAAAAATTAAAACGGAGTATGGTATGGATGGTAAAGATGCTAAGATCAAACGTAAAAATGCTACTAAAAGAAAATCAAATTCCAAAAAGATAGATAATCCAAAAAATAATTTAAAAATGATGTGCGAGGAATGCGGAATTACTGTAAAAGATTTAAAAGAGCATATGAGACAACATCTTCCTCCAGACGAGAGACCAAGAGTCAATTGTCACCTTTGTGAGGAAACGTTTTCATCGGCCAATTCAAAATACAAACATGTGAGACGTAAACATATGAACATCAAACAGAATTGTAAAATCTGTAATAAGGCTGTCGCAAACCTGAAAGCGCACATATTAGTGGTGCACAATACGGAATCACTGCCGTTTGAATGTATAGCATGCGGACGCGGGTTCATATCGAAGTCGCGCCTTGACGTTCATATGGCGGTGCACACCAAGGATCGCCCTCATAAATGCTCATTTTGCATTAAACGGTTTAGGACTAAGATTTGCATGCAGCTACACGAGCGACAAGTCCACAAAAAGGAGAAGAATCATCTCTGTCAGTTCTGTTCGAAAACTTTCTTCAAAAAATATCACTTACAAGTGCATATAAGAACTCACACGAAGGAGAAACCGTACGAATGCAACGAGTGCGGCAAGTGGTTCAGCTCGAAGAACGTGCTGAAGAACCACAAGCTCATACACGAGGACGTCAAGAGATTCGCGTGCACGCTCTGCGATATGTCGTTCATTAGAAACGGGTACCTGCATGCGCACATGCTGTCCCACAAGAAAGAGAAACGCTTCGAGTGTCAGTACTGCGGGGCGAAGTTCCACCGCTCCGACCACAGGAGGCGCCACGAGCACACCGTGCACCGCCGACACCTCGACAACGCTCACGACACACCCTTCACGGCCATATAA
Protein
MAGRMLKFIRGKGQQPSAERQKLQNELFAFRKTVQHGFPHRASALAWDPLLRLAALGTATGALKVYGRPGVELYGQHTNYESIVTQIHFITGTGRLISLCDDNSLHLWEINEKSLVELKSHVFEGKNKKISAICVESSAKNALLGTEGGNIYSLDLSAFTISEDVIYQDVVMQNCPEDYKVNPGAVESVYEHPKTPTRILIGYNRGLAVLWDREAAAPTHTFVSNQQLESLCWNDDGEHFTSSHNDGSYVTWEVAGASSDRPLKEPVTPYGPYPCKAITKILNRTSVDGEDITIFAGGMPRASYSDKYTVTVQQGEKHVAFDFTSRVIDLFTTTPVPPDGAPLQEEQPDTPALVQVQAVNQVAAALVVLAEEELVVIDLCDARWRPLRLPYLVSVHASAVTTAQLADGVAPAVYDNIVAAGQQQTENQYSESPWPISGGVVETAELTDRQVLLTGHEDGSVRFWDVTGVAMTPLYKYTTAQLFSGEELGENNDSQTTDEEEWPPFRRVGTFDPYSDDPRLAVKKVILCPLSGMLTIGGAAGHIVIASLKTSPSTTEVKSVSVNIVSDRDGFVWKGHGQLTLRAGTHTFPQGYQATSVVQLQPPAAVTSLSAQWEWGVVCAGTAHGLALVDALGPAPAPLLHKCTLNPHDHGSGDTPISRRKSFKKSLRESFRRLRKGRSQRRQTTSNPTSPTQPIPKKPIDKAASETDAEIKPIERAVEARSTDDAYGSMVRCLYFARTFLINTQQSTPTLWAGTNNGTVYAFTLHVPNTNKRKEEPVTCQLAKEIQLKHRAPVIGITVLDGASVPLPDPLEVERGVSPLPEPGTHRVVITSEEQFKIFTLPSLKPHNKYKLTAHEGARVRRTAWSQFSCGAHREWCLLCLTNLGDCLVLSPELRRQLNAAAVRKEDINGISSLCFSKRGEALYLHSSSELQRITLSATKVTIAECHVLLAPWAAALRGPAPEPPLTNGEHKVDCVLIVGLTTQEEVPETAHDVTAASADITVDSVRDHTSAEHPPELNINLQNTQVNTSSMVIKTTTRTTVNETNADGTTTTTTTTSTNNANENILEHSREEGVITRIETGTVTVPAGTDPKLILEMFDRQRSPLAVVKNPTITTKLCAKCYRKILSFHKFKSLAIENDIYLESQCTSAKHQIPLNNNENEIGPTQQFSIQMVDCLSPEMNNKDIIFREQVKTEMVLAHDEENDSIELGKSEPPELESDDEYLSVIKKIKTEYGMDGKDAKIKRKNATKRKSNSKKIDNPKNNLKMMCEECGITVKDLKEHMRQHLPPDERPRVNCHLCEETFSSANSKYKHVRRKHMNIKQNCKICNKAVANLKAHILVVHNTESLPFECIACGRGFISKSRLDVHMAVHTKDRPHKCSFCIKRFRTKICMQLHERQVHKKEKNHLCQFCSKTFFKKYHLQVHIRTHTKEKPYECNECGKWFSSKNVLKNHKLIHEDVKRFACTLCDMSFIRNGYLHAHMLSHKKEKRFECQYCGAKFHRSDHRRRHEHTVHRRHLDNAHDTPFTAI
Summary
Uniprot
A0A194QB38
A0A2J7Q3S4
A0A2J7Q3P7
A0A3L8DEL2
A0A195CTM4
A0A158NTR4
+ More
A0A0P4VYT6 A0A023F356 A0A069DZP9 A0A224X621 A0A0V0G3S9 T1HCU9 A0A1L8DVZ1 A0A1L8DW03 A0A1L8DW02 W5MCA3 Q6NRG4 A0A2Y9M323 A0A2Y9M5S3 A0A2Y9MGN2 F6Z9W6 A0A2Y9T6I3 A0A2Y9MBF5 A0A2K5CZU7 A0A1S3F7U4 A0A1S3F7A4 A0A2K5CZW4 E2RPS3 F7CIU8 A0A1S3F5K5 A0A0D9R738 A0A3Q7NGW2
A0A0P4VYT6 A0A023F356 A0A069DZP9 A0A224X621 A0A0V0G3S9 T1HCU9 A0A1L8DVZ1 A0A1L8DW03 A0A1L8DW02 W5MCA3 Q6NRG4 A0A2Y9M323 A0A2Y9M5S3 A0A2Y9MGN2 F6Z9W6 A0A2Y9T6I3 A0A2Y9MBF5 A0A2K5CZU7 A0A1S3F7U4 A0A1S3F7A4 A0A2K5CZW4 E2RPS3 F7CIU8 A0A1S3F5K5 A0A0D9R738 A0A3Q7NGW2
EMBL
KQ459232
KPJ02644.1
NEVH01019065
PNF23209.1
PNF23211.1
QOIP01000009
+ More
RLU18875.1 KQ977279 KYN04048.1 ADTU01026117 GDKW01001880 JAI54715.1 GBBI01003141 JAC15571.1 GBGD01000260 JAC88629.1 GFTR01008501 JAW07925.1 GECL01003493 JAP02631.1 ACPB03000213 GFDF01003477 JAV10607.1 GFDF01003478 JAV10606.1 GFDF01003476 JAV10608.1 AHAT01013466 AHAT01013467 AHAT01013468 AHAT01013469 BC070788 AAH70788.1 AAEX03003715 AQIB01148471 AQIB01148472
RLU18875.1 KQ977279 KYN04048.1 ADTU01026117 GDKW01001880 JAI54715.1 GBBI01003141 JAC15571.1 GBGD01000260 JAC88629.1 GFTR01008501 JAW07925.1 GECL01003493 JAP02631.1 ACPB03000213 GFDF01003477 JAV10607.1 GFDF01003478 JAV10606.1 GFDF01003476 JAV10608.1 AHAT01013466 AHAT01013467 AHAT01013468 AHAT01013469 BC070788 AAH70788.1 AAEX03003715 AQIB01148471 AQIB01148472
Proteomes
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
A0A194QB38
A0A2J7Q3S4
A0A2J7Q3P7
A0A3L8DEL2
A0A195CTM4
A0A158NTR4
+ More
A0A0P4VYT6 A0A023F356 A0A069DZP9 A0A224X621 A0A0V0G3S9 T1HCU9 A0A1L8DVZ1 A0A1L8DW03 A0A1L8DW02 W5MCA3 Q6NRG4 A0A2Y9M323 A0A2Y9M5S3 A0A2Y9MGN2 F6Z9W6 A0A2Y9T6I3 A0A2Y9MBF5 A0A2K5CZU7 A0A1S3F7U4 A0A1S3F7A4 A0A2K5CZW4 E2RPS3 F7CIU8 A0A1S3F5K5 A0A0D9R738 A0A3Q7NGW2
A0A0P4VYT6 A0A023F356 A0A069DZP9 A0A224X621 A0A0V0G3S9 T1HCU9 A0A1L8DVZ1 A0A1L8DW03 A0A1L8DW02 W5MCA3 Q6NRG4 A0A2Y9M323 A0A2Y9M5S3 A0A2Y9MGN2 F6Z9W6 A0A2Y9T6I3 A0A2Y9MBF5 A0A2K5CZU7 A0A1S3F7U4 A0A1S3F7A4 A0A2K5CZW4 E2RPS3 F7CIU8 A0A1S3F5K5 A0A0D9R738 A0A3Q7NGW2
PDB
6N8S
E-value=0,
Score=1805
Ontologies
GO
Topology
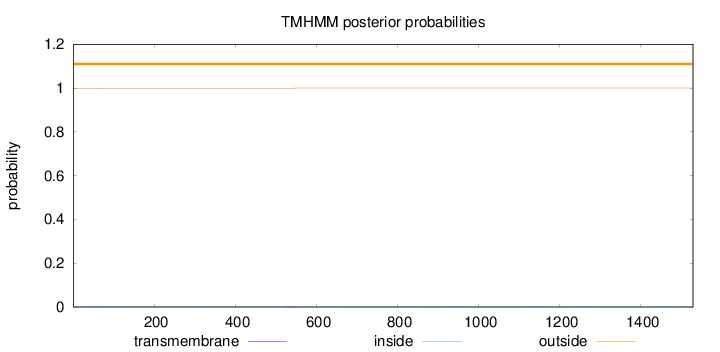
Length:
1529
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0422100000000001
Exp number, first 60 AAs:
0.00834
Total prob of N-in:
0.00163
outside
1 - 1529
Population Genetic Test Statistics
Pi
222.151838
Theta
154.503407
Tajima's D
0
CLR
11.069992
CSRT
0.371431428428579
Interpretation
Uncertain
