Gene
KWMTBOMO10153
Pre Gene Modal
BGIBMGA005567
Annotation
HM00051_[Heliconius_melpomene]
Location in the cell
Mitochondrial Reliability : 1.5 Nuclear Reliability : 1.43
Sequence
CDS
ATGCGTCGTCTGGAATTGAACTGGCTCCGGACTCGTTATTCAGAGTTACAGCGCGAACGACGTGGGCTGCATGCACAGATCCTACGTCTGAGATCTTTGTACACTCAACAACAACTACTATTGGAAAGTCTTTGGGGCAACGATCTCCGCCCGGGCACGGCGCAAGAGAGTGCCCTGACTAAGGCGTCGGCGTTACGTGACGCCTTAGCCTCTGTGAGTGCCAAGCTACGGGCGGCCGCGGAGTACGCACACGCTGCCGTCAGACTCCTCGATGATGCTCTGCCCGCTTGGAAACTCACCTCAGTCGGCAAGTGGGTGTCATAA
Protein
MRRLELNWLRTRYSELQRERRGLHAQILRLRSLYTQQQLLLESLWGNDLRPGTAQESALTKASALRDALASVSAKLRAAAEYAHAAVRLLDDALPAWKLTSVGKWVS
Summary
Similarity
Belongs to the complex I 75 kDa subunit family.
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF54292
SSF54292
CDD
ProteinModelPortal
Ontologies
KEGG
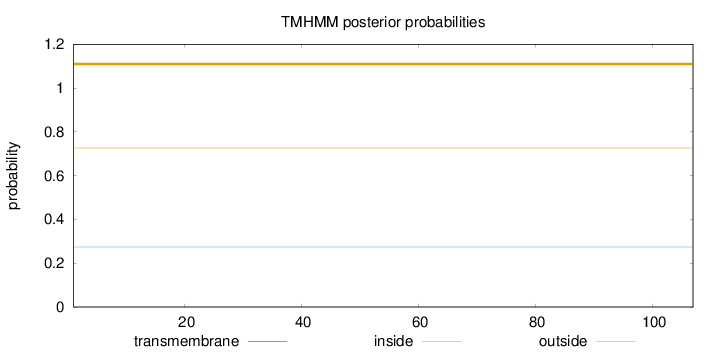
Topology
Length:
107
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00078
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.27401
outside
1 - 107
Population Genetic Test Statistics
Pi
154.10994
Theta
130.683268
Tajima's D
0.500777
CLR
0
CSRT
0.516874156292185
Interpretation
Uncertain
