Gene
KWMTBOMO10145
Annotation
PREDICTED:_INO80_complex_subunit_C_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 3.502
Sequence
CDS
ATGTCTAATGAATTAGAGAAATATTATTGTTTCAAGAGAGAAAACAAATTCATTGTAAAAAGTGGATCTAAAAAACGTATGTGGAGATCTTTAAAACAAATCTTAACAGCCGATCGAGCATTACCATGGCCAAATGAAGCAGTTTTATATTATTCAATAAATGCTCCGCCAACATTCAAGCCTATTAAAAAATATTCAGACATATCAGGATTACCAGCACCATATATGGATAGACACTCAAAACTTTATTTCTCTAATGCTGAAGAGTTTGCAACAGTCAGGAACCTTCCAATGGACATAACAGCAGGATATTTACAACTCAGAGGAGCAAACACTATCGTGGGATAA
Protein
MSNELEKYYCFKRENKFIVKSGSKKRMWRSLKQILTADRALPWPNEAVLYYSINAPPTFKPIKKYSDISGLPAPYMDRHSKLYFSNAEEFATVRNLPMDITAGYLQLRGANTIVG
Summary
Uniprot
A0A2A4ISE2
S4PPK9
L7X1P5
A0A2H1VL66
D0AB93
A0A1E1WTB0
+ More
A0A194QXJ5 A0A194QAZ5 H9J7S3 A0A212EP79 A0A154PBW8 A0A0K8TRM9 A0A0C9RK32 A0A0M8ZPT4 A0A0L7RDE6 A0A151IGU5 A0A232EQH8 A0A1L8DWG2 A0A158P345 E2AJK9 A0A151WFC5 A0A0J7KM36 A0A026WE77 E2BX55 A0A195BD12 F4WM73 A0A1B0ETW9 A0A195F3U5 A0A088ARQ6 A0A1Q3F8F2 B0W4J6 D6WC71 A0A1W4WB91 A0A1B0D5B6 A0A1B6DG18 A0A2A3E475 A0A0L0C1P4 A0A182H8P8 A0A1I8MEC0 A0A182U5L9 A0A182XFX3 A0NF19 A0A182IC58 Q17K42 A0A1Y1M3W8 A0A084WH65 U5EQC1 A0A2M4AXN5 T1GQ28 U4UG07 A0A1B6L365 A0A195DCF2 E9IEY2 A0A182F3G6 A0A2M4C3W4 A0A182RDS1 A0A2M4AQ96 A0A1B6G3S4 W5JTV3 A0A182YDT7 A0A182W6F1 W8C1P4 A0A2M3ZDN0 A0A182J0F7 A0A182NU04 A0A182QE30 A0A1A9W949 A0A2H8TS43 R4G879 A0A069DVU3 A0A023F9E0 N6UIN1 E0VXT3 T1HFC8 C4WS16 A0A224Y2Z0 A0A0P4VP97 A0A182PLS1 A0A1B0APP8 A0A1A9XXS7 S4RGB8 A0A0P4W6U2 A0A0K8V9R4 A0A067R655 A0A182K1H7 A0A1I8PPF9 A0A0K8SIZ7 A0A0A9Y6K6 A0A0V0GCH2 A0A093H4D1 A0A1A9VBL6 A0A091TRD2 A0A2P4T5K2 R0JIC3 A0A093HLA8 G1N4F7 A0A091NYD1 A0A093FSI6 A0A099ZDC5 A0A1A9Z5V1
A0A194QXJ5 A0A194QAZ5 H9J7S3 A0A212EP79 A0A154PBW8 A0A0K8TRM9 A0A0C9RK32 A0A0M8ZPT4 A0A0L7RDE6 A0A151IGU5 A0A232EQH8 A0A1L8DWG2 A0A158P345 E2AJK9 A0A151WFC5 A0A0J7KM36 A0A026WE77 E2BX55 A0A195BD12 F4WM73 A0A1B0ETW9 A0A195F3U5 A0A088ARQ6 A0A1Q3F8F2 B0W4J6 D6WC71 A0A1W4WB91 A0A1B0D5B6 A0A1B6DG18 A0A2A3E475 A0A0L0C1P4 A0A182H8P8 A0A1I8MEC0 A0A182U5L9 A0A182XFX3 A0NF19 A0A182IC58 Q17K42 A0A1Y1M3W8 A0A084WH65 U5EQC1 A0A2M4AXN5 T1GQ28 U4UG07 A0A1B6L365 A0A195DCF2 E9IEY2 A0A182F3G6 A0A2M4C3W4 A0A182RDS1 A0A2M4AQ96 A0A1B6G3S4 W5JTV3 A0A182YDT7 A0A182W6F1 W8C1P4 A0A2M3ZDN0 A0A182J0F7 A0A182NU04 A0A182QE30 A0A1A9W949 A0A2H8TS43 R4G879 A0A069DVU3 A0A023F9E0 N6UIN1 E0VXT3 T1HFC8 C4WS16 A0A224Y2Z0 A0A0P4VP97 A0A182PLS1 A0A1B0APP8 A0A1A9XXS7 S4RGB8 A0A0P4W6U2 A0A0K8V9R4 A0A067R655 A0A182K1H7 A0A1I8PPF9 A0A0K8SIZ7 A0A0A9Y6K6 A0A0V0GCH2 A0A093H4D1 A0A1A9VBL6 A0A091TRD2 A0A2P4T5K2 R0JIC3 A0A093HLA8 G1N4F7 A0A091NYD1 A0A093FSI6 A0A099ZDC5 A0A1A9Z5V1
Pubmed
23622113
23674305
26354079
19121390
22118469
26369729
+ More
28648823 21347285 20798317 24508170 30249741 21719571 18362917 19820115 26108605 26483478 25315136 12364791 14747013 17210077 17510324 28004739 24438588 23537049 21282665 20920257 23761445 25244985 24495485 26334808 25474469 20566863 27129103 24845553 25401762 20838655
28648823 21347285 20798317 24508170 30249741 21719571 18362917 19820115 26108605 26483478 25315136 12364791 14747013 17210077 17510324 28004739 24438588 23537049 21282665 20920257 23761445 25244985 24495485 26334808 25474469 20566863 27129103 24845553 25401762 20838655
EMBL
NWSH01007563
PCG62907.1
GAIX01000187
JAA92373.1
KC469893
AGC92698.1
+ More
ODYU01003152 SOQ41546.1 FP102340 CBH09288.1 GDQN01001013 JAT90041.1 KQ461155 KPJ08296.1 KQ459232 KPJ02637.1 BABH01020197 AGBW02013517 OWR43284.1 KQ434856 KZC08700.1 GDAI01001028 JAI16575.1 GBYB01008570 JAG78337.1 KQ438551 KOX67173.1 KQ414614 KOC68825.1 KQ977726 KYN00293.1 NNAY01002764 OXU20613.1 GFDF01003304 JAV10780.1 ADTU01001303 GL440027 EFN66404.1 KQ983219 KYQ46534.1 LBMM01005677 KMQ91296.1 KK107293 QOIP01000005 EZA53334.1 RLU23076.1 GL451202 EFN79774.1 KQ976522 KYM82084.1 GL888217 EGI64717.1 AJWK01010437 KQ981855 KYN34754.1 GFDL01011236 JAV23809.1 DS231838 EDS33841.1 KQ971316 EEZ98770.1 AJVK01011728 GEDC01012681 JAS24617.1 KZ288427 PBC25881.1 JRES01001123 KNC25339.1 JXUM01118902 KQ566235 KXJ70297.1 AAAB01008964 EAU76314.1 APCN01000775 CH477228 EAT47050.1 GEZM01041321 GEZM01041320 JAV80512.1 ATLV01023791 KE525346 KFB49559.1 GANO01004321 JAB55550.1 GGFK01012248 MBW45569.1 CAQQ02031859 KB632308 ERL91957.1 GEBQ01021849 JAT18128.1 KQ980989 KYN10563.1 GL762728 EFZ20875.1 GGFJ01010876 MBW60017.1 GGFK01009633 MBW42954.1 GECZ01012680 JAS57089.1 ADMH02000463 ETN66380.1 GAMC01000824 GAMC01000823 JAC05733.1 GGFM01005840 MBW26591.1 AXCN02000808 GFXV01005220 MBW17025.1 GAHY01001442 JAA76068.1 GBGD01003475 JAC85414.1 GBBI01000596 JAC18116.1 APGK01023763 KB740512 ENN80511.1 DS235840 EEB18189.1 ACPB03021649 ABLF02029934 AK340009 BAH70686.1 GFTR01001555 JAW14871.1 GDKW01002296 JAI54299.1 JXJN01001542 GDRN01067655 JAI64337.1 GDHF01016746 JAI35568.1 KK852675 KDR18718.1 GBRD01013057 GBRD01013056 JAG52770.1 GBHO01016323 GBHO01016322 JAG27281.1 JAG27282.1 GECL01000201 JAP05923.1 KL205937 KFV77423.1 KK463913 KFQ80703.1 PPHD01008162 POI31604.1 KB743886 EOA96751.1 KK630440 KFV55388.1 KK644595 KFP98056.1 KK400496 KFV59645.1 KL893134 KGL80414.1
ODYU01003152 SOQ41546.1 FP102340 CBH09288.1 GDQN01001013 JAT90041.1 KQ461155 KPJ08296.1 KQ459232 KPJ02637.1 BABH01020197 AGBW02013517 OWR43284.1 KQ434856 KZC08700.1 GDAI01001028 JAI16575.1 GBYB01008570 JAG78337.1 KQ438551 KOX67173.1 KQ414614 KOC68825.1 KQ977726 KYN00293.1 NNAY01002764 OXU20613.1 GFDF01003304 JAV10780.1 ADTU01001303 GL440027 EFN66404.1 KQ983219 KYQ46534.1 LBMM01005677 KMQ91296.1 KK107293 QOIP01000005 EZA53334.1 RLU23076.1 GL451202 EFN79774.1 KQ976522 KYM82084.1 GL888217 EGI64717.1 AJWK01010437 KQ981855 KYN34754.1 GFDL01011236 JAV23809.1 DS231838 EDS33841.1 KQ971316 EEZ98770.1 AJVK01011728 GEDC01012681 JAS24617.1 KZ288427 PBC25881.1 JRES01001123 KNC25339.1 JXUM01118902 KQ566235 KXJ70297.1 AAAB01008964 EAU76314.1 APCN01000775 CH477228 EAT47050.1 GEZM01041321 GEZM01041320 JAV80512.1 ATLV01023791 KE525346 KFB49559.1 GANO01004321 JAB55550.1 GGFK01012248 MBW45569.1 CAQQ02031859 KB632308 ERL91957.1 GEBQ01021849 JAT18128.1 KQ980989 KYN10563.1 GL762728 EFZ20875.1 GGFJ01010876 MBW60017.1 GGFK01009633 MBW42954.1 GECZ01012680 JAS57089.1 ADMH02000463 ETN66380.1 GAMC01000824 GAMC01000823 JAC05733.1 GGFM01005840 MBW26591.1 AXCN02000808 GFXV01005220 MBW17025.1 GAHY01001442 JAA76068.1 GBGD01003475 JAC85414.1 GBBI01000596 JAC18116.1 APGK01023763 KB740512 ENN80511.1 DS235840 EEB18189.1 ACPB03021649 ABLF02029934 AK340009 BAH70686.1 GFTR01001555 JAW14871.1 GDKW01002296 JAI54299.1 JXJN01001542 GDRN01067655 JAI64337.1 GDHF01016746 JAI35568.1 KK852675 KDR18718.1 GBRD01013057 GBRD01013056 JAG52770.1 GBHO01016323 GBHO01016322 JAG27281.1 JAG27282.1 GECL01000201 JAP05923.1 KL205937 KFV77423.1 KK463913 KFQ80703.1 PPHD01008162 POI31604.1 KB743886 EOA96751.1 KK630440 KFV55388.1 KK644595 KFP98056.1 KK400496 KFV59645.1 KL893134 KGL80414.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000005204
UP000007151
UP000076502
+ More
UP000053105 UP000053825 UP000078542 UP000215335 UP000005205 UP000000311 UP000075809 UP000036403 UP000053097 UP000279307 UP000008237 UP000078540 UP000007755 UP000092461 UP000078541 UP000005203 UP000002320 UP000007266 UP000192223 UP000092462 UP000242457 UP000037069 UP000069940 UP000249989 UP000095301 UP000075902 UP000076407 UP000007062 UP000075840 UP000008820 UP000030765 UP000015102 UP000030742 UP000078492 UP000069272 UP000075900 UP000000673 UP000076408 UP000075920 UP000075880 UP000075884 UP000075886 UP000091820 UP000019118 UP000009046 UP000015103 UP000007819 UP000075885 UP000092460 UP000092443 UP000245300 UP000027135 UP000075881 UP000095300 UP000053584 UP000078200 UP000001645 UP000053641 UP000092445
UP000053105 UP000053825 UP000078542 UP000215335 UP000005205 UP000000311 UP000075809 UP000036403 UP000053097 UP000279307 UP000008237 UP000078540 UP000007755 UP000092461 UP000078541 UP000005203 UP000002320 UP000007266 UP000192223 UP000092462 UP000242457 UP000037069 UP000069940 UP000249989 UP000095301 UP000075902 UP000076407 UP000007062 UP000075840 UP000008820 UP000030765 UP000015102 UP000030742 UP000078492 UP000069272 UP000075900 UP000000673 UP000076408 UP000075920 UP000075880 UP000075884 UP000075886 UP000091820 UP000019118 UP000009046 UP000015103 UP000007819 UP000075885 UP000092460 UP000092443 UP000245300 UP000027135 UP000075881 UP000095300 UP000053584 UP000078200 UP000001645 UP000053641 UP000092445
PRIDE
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
ProteinModelPortal
A0A2A4ISE2
S4PPK9
L7X1P5
A0A2H1VL66
D0AB93
A0A1E1WTB0
+ More
A0A194QXJ5 A0A194QAZ5 H9J7S3 A0A212EP79 A0A154PBW8 A0A0K8TRM9 A0A0C9RK32 A0A0M8ZPT4 A0A0L7RDE6 A0A151IGU5 A0A232EQH8 A0A1L8DWG2 A0A158P345 E2AJK9 A0A151WFC5 A0A0J7KM36 A0A026WE77 E2BX55 A0A195BD12 F4WM73 A0A1B0ETW9 A0A195F3U5 A0A088ARQ6 A0A1Q3F8F2 B0W4J6 D6WC71 A0A1W4WB91 A0A1B0D5B6 A0A1B6DG18 A0A2A3E475 A0A0L0C1P4 A0A182H8P8 A0A1I8MEC0 A0A182U5L9 A0A182XFX3 A0NF19 A0A182IC58 Q17K42 A0A1Y1M3W8 A0A084WH65 U5EQC1 A0A2M4AXN5 T1GQ28 U4UG07 A0A1B6L365 A0A195DCF2 E9IEY2 A0A182F3G6 A0A2M4C3W4 A0A182RDS1 A0A2M4AQ96 A0A1B6G3S4 W5JTV3 A0A182YDT7 A0A182W6F1 W8C1P4 A0A2M3ZDN0 A0A182J0F7 A0A182NU04 A0A182QE30 A0A1A9W949 A0A2H8TS43 R4G879 A0A069DVU3 A0A023F9E0 N6UIN1 E0VXT3 T1HFC8 C4WS16 A0A224Y2Z0 A0A0P4VP97 A0A182PLS1 A0A1B0APP8 A0A1A9XXS7 S4RGB8 A0A0P4W6U2 A0A0K8V9R4 A0A067R655 A0A182K1H7 A0A1I8PPF9 A0A0K8SIZ7 A0A0A9Y6K6 A0A0V0GCH2 A0A093H4D1 A0A1A9VBL6 A0A091TRD2 A0A2P4T5K2 R0JIC3 A0A093HLA8 G1N4F7 A0A091NYD1 A0A093FSI6 A0A099ZDC5 A0A1A9Z5V1
A0A194QXJ5 A0A194QAZ5 H9J7S3 A0A212EP79 A0A154PBW8 A0A0K8TRM9 A0A0C9RK32 A0A0M8ZPT4 A0A0L7RDE6 A0A151IGU5 A0A232EQH8 A0A1L8DWG2 A0A158P345 E2AJK9 A0A151WFC5 A0A0J7KM36 A0A026WE77 E2BX55 A0A195BD12 F4WM73 A0A1B0ETW9 A0A195F3U5 A0A088ARQ6 A0A1Q3F8F2 B0W4J6 D6WC71 A0A1W4WB91 A0A1B0D5B6 A0A1B6DG18 A0A2A3E475 A0A0L0C1P4 A0A182H8P8 A0A1I8MEC0 A0A182U5L9 A0A182XFX3 A0NF19 A0A182IC58 Q17K42 A0A1Y1M3W8 A0A084WH65 U5EQC1 A0A2M4AXN5 T1GQ28 U4UG07 A0A1B6L365 A0A195DCF2 E9IEY2 A0A182F3G6 A0A2M4C3W4 A0A182RDS1 A0A2M4AQ96 A0A1B6G3S4 W5JTV3 A0A182YDT7 A0A182W6F1 W8C1P4 A0A2M3ZDN0 A0A182J0F7 A0A182NU04 A0A182QE30 A0A1A9W949 A0A2H8TS43 R4G879 A0A069DVU3 A0A023F9E0 N6UIN1 E0VXT3 T1HFC8 C4WS16 A0A224Y2Z0 A0A0P4VP97 A0A182PLS1 A0A1B0APP8 A0A1A9XXS7 S4RGB8 A0A0P4W6U2 A0A0K8V9R4 A0A067R655 A0A182K1H7 A0A1I8PPF9 A0A0K8SIZ7 A0A0A9Y6K6 A0A0V0GCH2 A0A093H4D1 A0A1A9VBL6 A0A091TRD2 A0A2P4T5K2 R0JIC3 A0A093HLA8 G1N4F7 A0A091NYD1 A0A093FSI6 A0A099ZDC5 A0A1A9Z5V1
PDB
6FML
E-value=8.16151e-12,
Score=163
Ontologies
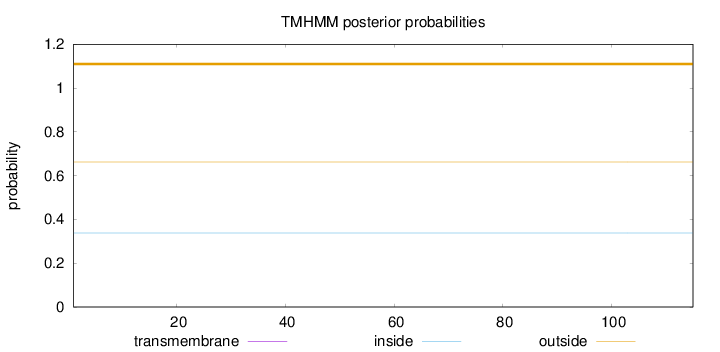
Topology
Length:
115
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00369
Exp number, first 60 AAs:
0.00204
Total prob of N-in:
0.33801
outside
1 - 115
Population Genetic Test Statistics
Pi
126.055729
Theta
82.520273
Tajima's D
1.651393
CLR
0
CSRT
0.821508924553772
Interpretation
Uncertain
