Pre Gene Modal
BGIBMGA005637
Annotation
Protein_penguin_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.643
Sequence
CDS
ATGCAGAAAGGTAAAAGGAAAAGTGATGAAGATGGTGTTAGTTCACCAAAAAAGAAAAAACTAAATAGTTCGCATGAACAAGATAATAAAGTTAAAAACATCAAAAACAAAATAAACCTAAAAGGTAAAAAATTTAATAAGGATGGAGTAAAAGGTTTCAAAAAATTTACTAAAAACGACAAAGGATCTAAATTTGTTAAAACAAAAGAAAAATCGAATTTTTCTAAAGATGAAGCAAACGGAGAAAAGCCAAAATGGGCTGAAATGAAAAAAGAGAAAAAACAACTAAGGACTGTCAGACGTAAAGCCAAAATAACAGCTGAAGTATTTGAGATTTCACATAAGGCCAAGTTGTTAGCAGCACAAATACAGAGAAAAGTTGTGAAATCTGACTTCAAAATAAACTCCTGTAAAGAATTGCACAATATGTTAAAGGGACAATACAAGTCTATTGCTCTCACACATGATCTTAGTCGAGTTATTCAAGTTCTACTAAAACATAGCCCTGAAGACATTATAAATGAAATAACGAAAGAGCTTCTAGACATTATAGTACAGATGGGTCAATCTAAGTACGCGCATCATTCCGTTAAGCGAATACTGAAATATGGAACAGATTTCATTCGGCATGAGGTTTTGAAGAAGTTTTATGGACATGTTGTATCCCTGTCAACTCATGCTATTAGTGCACCTGTCTTGGATTATGCATATGGGGAGTTTGCCTCGAAAAAGGAAAAATGCCACATGCAACAAGAATTCTATGGAGAGATTTATAAGAATACAAAAGACGACAAGGTAAAGACACTCAGAGATGCATACAAAGACAGTCCAGAAATGAAATCAGCTATACTCCAGGCCTGTAAGACCAACTTACAGAAGATATTAGACAAAAACCTACATGATGGAGAATTGTTTCATTCTGTGTTATATGACTATATCTGTGAATGCAGCCAAGAAGATAGAGTAGATTTTATTTCTATTCTTAGCCCACTGATTGTCCCACTCAGTAATTCTTTACCTGGTGTCAATGCTGCCAGTATGTGCGTTTGGCAGGGTACAAATAAGGACAAGAAAACAATTTTAAAAGTGGTCAAGGAACATGCTATTCCTTTAAAAGACTATTGTGTCCACTCTGGCTGCCAATATAAAGACATTGCTAAGGACCACTGGGGTAATATGGTAATTCACTGGTTAGTAAAACCAAAAGACACAGCTGCTTTCCATCCTAGTTTAATTAAGTTTTTGGAAGATGGTGCTAAGAGTGGAACATCGAAGAAAGACACAGAAATTCGTGTTTTGGAGCTTAGGGAAGCTATATTACCTGCTCTAAAGAAAGATATGGAAACAGATCCTAAATTCTGGCTAAGCAATAAATCTAATATGTTGTTGGCTGTTGCTGTATTGTCTATAGAATCATCTAAAGAGATTCTGCATGCATTGGCAAAAGTGATCTGCGAGACTGACTGGCACATGCAGGTGGAAGGGAAGGACGATACCCTGGCGATTGAGGATGCTGGCATACACATGTGCTTAAAAAAACTAGCCAGTTTAGACAAAGAAAAATCGGCTGATACTCTTGGTGAAGCTATAAGTGAACATTTAAATGATGAAACTATTAAAGTGTGGATACGTACAAATAGGGGTTGCTTTTTCCTATTGAAATTGATAGAGAGTAATCGTGAAGAAGTAGCTAAGAAATTAAAAGCAAAATTAAAACCACATGTAAATATTTTGAAGCAACAATCATCGGAAGGTGCTAAATTATTGTTACAGAATGTACAGAATAAATAG
Protein
MQKGKRKSDEDGVSSPKKKKLNSSHEQDNKVKNIKNKINLKGKKFNKDGVKGFKKFTKNDKGSKFVKTKEKSNFSKDEANGEKPKWAEMKKEKKQLRTVRRKAKITAEVFEISHKAKLLAAQIQRKVVKSDFKINSCKELHNMLKGQYKSIALTHDLSRVIQVLLKHSPEDIINEITKELLDIIVQMGQSKYAHHSVKRILKYGTDFIRHEVLKKFYGHVVSLSTHAISAPVLDYAYGEFASKKEKCHMQQEFYGEIYKNTKDDKVKTLRDAYKDSPEMKSAILQACKTNLQKILDKNLHDGELFHSVLYDYICECSQEDRVDFISILSPLIVPLSNSLPGVNAASMCVWQGTNKDKKTILKVVKEHAIPLKDYCVHSGCQYKDIAKDHWGNMVIHWLVKPKDTAAFHPSLIKFLEDGAKSGTSKKDTEIRVLELREAILPALKKDMETDPKFWLSNKSNMLLAVAVLSIESSKEILHALAKVICETDWHMQVEGKDDTLAIEDAGIHMCLKKLASLDKEKSADTLGEAISEHLNDETIKVWIRTNRGCFFLLKLIESNREEVAKKLKAKLKPHVNILKQQSSEGAKLLLQNVQNK
Summary
Uniprot
A0A2A4JDV9
A0A2H1VM78
A0A194QAR9
A0A194QTH1
D0AB89
L7X0R1
+ More
S4PGK9 A0A212EP73 A0A0L7LL06 D6WZ15 A0A0T6B3C6 N6U5R4 A0A1Y1MBF6 A0A232EX65 K7J9N0 E2BXL1 V9IFZ9 A0A2A3E3U3 A0A088ALU9 A0A0J7K4J0 A0A154PGZ6 E9IFM9 E2A7S4 A0A0K8USZ1 A0A195DCV3 F4WJT7 A0A3L8DD65 A0A026W9A4 A0A034VQG9 A0A195BCC0 A0A158NS35 A0A195FXT3 A0A2J7QKP0 U5EHI9 A0A1A9WZ92 A0A151WGY7 A0A1B0AIX9 A0A336MGJ9 A0A336MJ44 A0A3L8DD77 W8BH01 A0A067QIM4 A0A1A9UHS6 A0A195BYX8 A0A1L8DGJ9 A0A1B0D4D4 A0A1B0CMH0 A0A1J1J034 A0A0K8WGW0 A0A182H6R2 A0A1I8M975 T1P9Y9 A0A2J7QKP7 A0A1B0G1S7 A0A1I8Q8R4 A0A1S4FNX6 A0A0L0C855 Q16UF7 A0A3Q0IPH3 B4NCP2 A0A1W4VII6 B4JP18 B4MCX2 A0A1S3JCA0 A0A1S3JC21 Q29H32 A0A1S3K9S0 D0QWE7 A0A3B0K6S4 A0A3B0KVM8 A0A1B6L0Z9 A0A2C9JHF8 A0A1Q3EYU5 E9G064 A0A0P5W3J1 A0A0P4YL48 A0A0P6BTP4 A0A0P4ZN88 A0A164U8Z5 A0A0P5SBG7 A0A0P6ERM2
S4PGK9 A0A212EP73 A0A0L7LL06 D6WZ15 A0A0T6B3C6 N6U5R4 A0A1Y1MBF6 A0A232EX65 K7J9N0 E2BXL1 V9IFZ9 A0A2A3E3U3 A0A088ALU9 A0A0J7K4J0 A0A154PGZ6 E9IFM9 E2A7S4 A0A0K8USZ1 A0A195DCV3 F4WJT7 A0A3L8DD65 A0A026W9A4 A0A034VQG9 A0A195BCC0 A0A158NS35 A0A195FXT3 A0A2J7QKP0 U5EHI9 A0A1A9WZ92 A0A151WGY7 A0A1B0AIX9 A0A336MGJ9 A0A336MJ44 A0A3L8DD77 W8BH01 A0A067QIM4 A0A1A9UHS6 A0A195BYX8 A0A1L8DGJ9 A0A1B0D4D4 A0A1B0CMH0 A0A1J1J034 A0A0K8WGW0 A0A182H6R2 A0A1I8M975 T1P9Y9 A0A2J7QKP7 A0A1B0G1S7 A0A1I8Q8R4 A0A1S4FNX6 A0A0L0C855 Q16UF7 A0A3Q0IPH3 B4NCP2 A0A1W4VII6 B4JP18 B4MCX2 A0A1S3JCA0 A0A1S3JC21 Q29H32 A0A1S3K9S0 D0QWE7 A0A3B0K6S4 A0A3B0KVM8 A0A1B6L0Z9 A0A2C9JHF8 A0A1Q3EYU5 E9G064 A0A0P5W3J1 A0A0P4YL48 A0A0P6BTP4 A0A0P4ZN88 A0A164U8Z5 A0A0P5SBG7 A0A0P6ERM2
Pubmed
EMBL
NWSH01001832
PCG69966.1
ODYU01003152
SOQ41542.1
KQ459232
KPJ02633.1
+ More
KQ461155 KPJ08300.1 FP102340 CBH09284.1 KC469893 AGC92694.1 GAIX01003617 JAA88943.1 AGBW02013517 OWR43288.1 JTDY01000686 KOB76223.1 KQ971363 EFA09038.1 LJIG01016117 KRT81565.1 APGK01041594 KB740994 KB632255 ENN75986.1 ERL90654.1 GEZM01035601 JAV83159.1 NNAY01001792 OXU22907.1 AAZX01002711 GL451265 EFN79580.1 JR041372 AEY59426.1 KZ288391 PBC26358.1 LBMM01014327 KMQ85219.1 KQ434896 KZC10744.1 GL762880 EFZ20617.1 GL437382 EFN70542.1 GDHF01022839 JAI29475.1 KQ980989 KYN10708.1 GL888186 EGI65579.1 QOIP01000010 RLU18102.1 KK107374 EZA51594.1 GAKP01015149 GAKP01015145 GAKP01015141 JAC43803.1 KQ976528 KYM81867.1 ADTU01024454 KQ981208 KYN44654.1 NEVH01013260 PNF29156.1 GANO01003004 JAB56867.1 KQ983136 KYQ47082.1 UFQT01001175 SSX29325.1 UFQT01001201 SSX29481.1 RLU18103.1 GAMC01010297 GAMC01010295 JAB96258.1 KK853308 KDR08684.1 KQ978457 KYM93817.1 GFDF01008496 JAV05588.1 AJVK01011378 AJVK01011379 AJWK01018717 CVRI01000066 CRL05875.1 GDHF01002204 JAI50110.1 JXUM01114779 JXUM01114780 JXUM01114781 JXUM01114782 JXUM01114783 JXUM01114784 KQ565817 KXJ70629.1 KA644763 AFP59392.1 PNF29155.1 CCAG010007360 JRES01000777 KNC28412.1 CH477622 EAT38180.1 CH964239 EDW82601.2 CH916371 EDV92461.1 CH940660 EDW58044.1 CH379064 EAL31927.2 FJ821026 ACN94646.1 OUUW01000039 SPP89917.1 SPP89916.1 GEBQ01022642 JAT17335.1 GFDL01014579 JAV20466.1 GL732528 EFX87453.1 GDIP01092481 JAM11234.1 GDIP01227270 JAI96131.1 GDIP01025106 JAM78609.1 GDIP01210276 JAJ13126.1 LRGB01001581 KZS11152.1 GDIQ01094329 JAL57397.1 GDIQ01059302 JAN35435.1
KQ461155 KPJ08300.1 FP102340 CBH09284.1 KC469893 AGC92694.1 GAIX01003617 JAA88943.1 AGBW02013517 OWR43288.1 JTDY01000686 KOB76223.1 KQ971363 EFA09038.1 LJIG01016117 KRT81565.1 APGK01041594 KB740994 KB632255 ENN75986.1 ERL90654.1 GEZM01035601 JAV83159.1 NNAY01001792 OXU22907.1 AAZX01002711 GL451265 EFN79580.1 JR041372 AEY59426.1 KZ288391 PBC26358.1 LBMM01014327 KMQ85219.1 KQ434896 KZC10744.1 GL762880 EFZ20617.1 GL437382 EFN70542.1 GDHF01022839 JAI29475.1 KQ980989 KYN10708.1 GL888186 EGI65579.1 QOIP01000010 RLU18102.1 KK107374 EZA51594.1 GAKP01015149 GAKP01015145 GAKP01015141 JAC43803.1 KQ976528 KYM81867.1 ADTU01024454 KQ981208 KYN44654.1 NEVH01013260 PNF29156.1 GANO01003004 JAB56867.1 KQ983136 KYQ47082.1 UFQT01001175 SSX29325.1 UFQT01001201 SSX29481.1 RLU18103.1 GAMC01010297 GAMC01010295 JAB96258.1 KK853308 KDR08684.1 KQ978457 KYM93817.1 GFDF01008496 JAV05588.1 AJVK01011378 AJVK01011379 AJWK01018717 CVRI01000066 CRL05875.1 GDHF01002204 JAI50110.1 JXUM01114779 JXUM01114780 JXUM01114781 JXUM01114782 JXUM01114783 JXUM01114784 KQ565817 KXJ70629.1 KA644763 AFP59392.1 PNF29155.1 CCAG010007360 JRES01000777 KNC28412.1 CH477622 EAT38180.1 CH964239 EDW82601.2 CH916371 EDV92461.1 CH940660 EDW58044.1 CH379064 EAL31927.2 FJ821026 ACN94646.1 OUUW01000039 SPP89917.1 SPP89916.1 GEBQ01022642 JAT17335.1 GFDL01014579 JAV20466.1 GL732528 EFX87453.1 GDIP01092481 JAM11234.1 GDIP01227270 JAI96131.1 GDIP01025106 JAM78609.1 GDIP01210276 JAJ13126.1 LRGB01001581 KZS11152.1 GDIQ01094329 JAL57397.1 GDIQ01059302 JAN35435.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
UP000007266
+ More
UP000019118 UP000030742 UP000215335 UP000002358 UP000008237 UP000242457 UP000005203 UP000036403 UP000076502 UP000000311 UP000078492 UP000007755 UP000279307 UP000053097 UP000078540 UP000005205 UP000078541 UP000235965 UP000091820 UP000075809 UP000092445 UP000027135 UP000078200 UP000078542 UP000092462 UP000092461 UP000183832 UP000069940 UP000249989 UP000095301 UP000092444 UP000095300 UP000037069 UP000008820 UP000079169 UP000007798 UP000192221 UP000001070 UP000008792 UP000085678 UP000001819 UP000268350 UP000076420 UP000000305 UP000076858
UP000019118 UP000030742 UP000215335 UP000002358 UP000008237 UP000242457 UP000005203 UP000036403 UP000076502 UP000000311 UP000078492 UP000007755 UP000279307 UP000053097 UP000078540 UP000005205 UP000078541 UP000235965 UP000091820 UP000075809 UP000092445 UP000027135 UP000078200 UP000078542 UP000092462 UP000092461 UP000183832 UP000069940 UP000249989 UP000095301 UP000092444 UP000095300 UP000037069 UP000008820 UP000079169 UP000007798 UP000192221 UP000001070 UP000008792 UP000085678 UP000001819 UP000268350 UP000076420 UP000000305 UP000076858
Pfam
PF08144 CPL
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2A4JDV9
A0A2H1VM78
A0A194QAR9
A0A194QTH1
D0AB89
L7X0R1
+ More
S4PGK9 A0A212EP73 A0A0L7LL06 D6WZ15 A0A0T6B3C6 N6U5R4 A0A1Y1MBF6 A0A232EX65 K7J9N0 E2BXL1 V9IFZ9 A0A2A3E3U3 A0A088ALU9 A0A0J7K4J0 A0A154PGZ6 E9IFM9 E2A7S4 A0A0K8USZ1 A0A195DCV3 F4WJT7 A0A3L8DD65 A0A026W9A4 A0A034VQG9 A0A195BCC0 A0A158NS35 A0A195FXT3 A0A2J7QKP0 U5EHI9 A0A1A9WZ92 A0A151WGY7 A0A1B0AIX9 A0A336MGJ9 A0A336MJ44 A0A3L8DD77 W8BH01 A0A067QIM4 A0A1A9UHS6 A0A195BYX8 A0A1L8DGJ9 A0A1B0D4D4 A0A1B0CMH0 A0A1J1J034 A0A0K8WGW0 A0A182H6R2 A0A1I8M975 T1P9Y9 A0A2J7QKP7 A0A1B0G1S7 A0A1I8Q8R4 A0A1S4FNX6 A0A0L0C855 Q16UF7 A0A3Q0IPH3 B4NCP2 A0A1W4VII6 B4JP18 B4MCX2 A0A1S3JCA0 A0A1S3JC21 Q29H32 A0A1S3K9S0 D0QWE7 A0A3B0K6S4 A0A3B0KVM8 A0A1B6L0Z9 A0A2C9JHF8 A0A1Q3EYU5 E9G064 A0A0P5W3J1 A0A0P4YL48 A0A0P6BTP4 A0A0P4ZN88 A0A164U8Z5 A0A0P5SBG7 A0A0P6ERM2
S4PGK9 A0A212EP73 A0A0L7LL06 D6WZ15 A0A0T6B3C6 N6U5R4 A0A1Y1MBF6 A0A232EX65 K7J9N0 E2BXL1 V9IFZ9 A0A2A3E3U3 A0A088ALU9 A0A0J7K4J0 A0A154PGZ6 E9IFM9 E2A7S4 A0A0K8USZ1 A0A195DCV3 F4WJT7 A0A3L8DD65 A0A026W9A4 A0A034VQG9 A0A195BCC0 A0A158NS35 A0A195FXT3 A0A2J7QKP0 U5EHI9 A0A1A9WZ92 A0A151WGY7 A0A1B0AIX9 A0A336MGJ9 A0A336MJ44 A0A3L8DD77 W8BH01 A0A067QIM4 A0A1A9UHS6 A0A195BYX8 A0A1L8DGJ9 A0A1B0D4D4 A0A1B0CMH0 A0A1J1J034 A0A0K8WGW0 A0A182H6R2 A0A1I8M975 T1P9Y9 A0A2J7QKP7 A0A1B0G1S7 A0A1I8Q8R4 A0A1S4FNX6 A0A0L0C855 Q16UF7 A0A3Q0IPH3 B4NCP2 A0A1W4VII6 B4JP18 B4MCX2 A0A1S3JCA0 A0A1S3JC21 Q29H32 A0A1S3K9S0 D0QWE7 A0A3B0K6S4 A0A3B0KVM8 A0A1B6L0Z9 A0A2C9JHF8 A0A1Q3EYU5 E9G064 A0A0P5W3J1 A0A0P4YL48 A0A0P6BTP4 A0A0P4ZN88 A0A164U8Z5 A0A0P5SBG7 A0A0P6ERM2
PDB
4WZW
E-value=7.74832e-39,
Score=405
Ontologies
GO
PANTHER
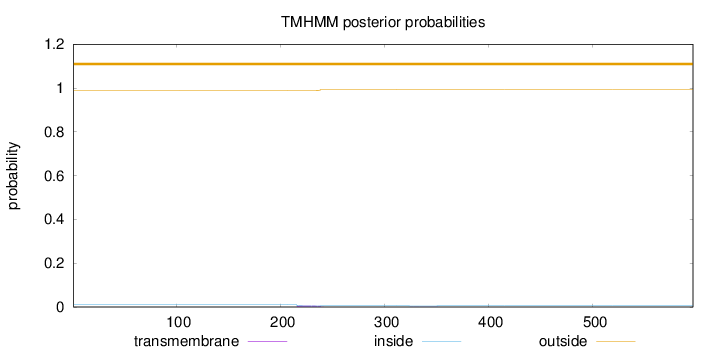
Topology
Length:
596
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15473
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01148
outside
1 - 596
Population Genetic Test Statistics
Pi
340.212503
Theta
172.649669
Tajima's D
2.881752
CLR
0.000821
CSRT
0.974401279936003
Interpretation
Uncertain
