Gene
KWMTBOMO10142
Pre Gene Modal
BGIBMGA005639
Annotation
PREDICTED:_DNA_fragmentation_factor_subunit_beta_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.729
Sequence
CDS
ATGAAGAAAGGTTATAAAGTGACTGATGTTAAACGCGAAAAGAAAATTGGCGTGGCAGCTGAAAGTTTACACGAGTTGATCGAGAAATCACACAAAAAATTAGAATTCAATGTCAGCTGCGGAGAATGTAATTTATACGTGGCTGAAGATGGGACGCTAGTTGACAATGATGAGTACCTAGCCACTCTGCCACCACAAACCGTCTTCATTTTACTAAAGGGCTCAGAAAAGATGATAACAGATTTTGATTATTATTACAATATGATCCTGTCAACTAAGAAAGAGTACATAGATACAGGAGCAGCAGCCAAGCAGTTTTTATCGACAAACATCAAAGAGAAGTTCAAAGTCTTTCAAAAGTACATTGCAGCTGCTGACGATGCTAGAATAGAGCTCAGCGAGAGAACACAAGATCCAGCTTGGTTTGAAGGTATAATGAAGATTTCCTTATACTTCATTTATTAA
Protein
MKKGYKVTDVKREKKIGVAAESLHELIEKSHKKLEFNVSCGECNLYVAEDGTLVDNDEYLATLPPQTVFILLKGSEKMITDFDYYYNMILSTKKEYIDTGAAAKQFLSTNIKEKFKVFQKYIAAADDARIELSERTQDPAWFEGIMKISLYFIY
Summary
Uniprot
H9J7Z7
A0A2H1VMR1
A0A1E1WJU4
A0A194QCC5
A0A194QS01
D0AB91
+ More
A0A212EP59 L7X1G4 A0A0L7LGR6 A0A0M9A9K2 V9IAB8 A0A310SHW6 W5JGP2 E0VAJ8 A0A1B6D4X6 A0A1B6CDC6 A0A182NBH5 A0A154PA95 A0A2J7PXZ6 A0A195FWH6 A0A182J889 A0A1B6EA14 A0A182QKM7 A0A182M0H1 A0A182VTZ2 A0A158NN72 A0A2A3EK54 A0A026VZ81 A0A0C9QV11 A0A084VH37 A0A3L8DG10 A0A182R3W6 A0A067QYQ3 F4WNU5 K7J7X5 A0A182K9P8 A0A182F0Q3 A0A182YFV0 A0A2P8ZLG4 A0A182PM90 A0A1B6FSK2 A0A146LJ53 Q7Q1Q3 A0A1B0ETL7 A0A182U7Z1 A0A146LWA4 A0A182XN40 A0A182HI82 N6SU96 U4U402 A0A232FLH6 A0A0T6AZL6 D6WW09 B0WHX2 A0A088ARL5 U5EPW9 Q17BG6 A0A1S4F990 A0A1B6KTR6 A0A182GGF6 A0A023F0S3 A0A224XFN9 A0A0P4VIT5 A0A1Y1KMX5 A0A069DSR6 A0A0V0GEA2 A0A2S2QP19 A0A1Y1KI19 J9KG38 A0A336MRH6 A0A182VLF9 A0A182LM80 A0A3M6UJD4 A0A210QD94 A0A0K8UQ57 A0A034WHR1 K1P4N7 A0A195CRW1 A0A2B4RSP0 A0A0B7BH20 B3MV75 B3NAZ4 B4M987 A0A1W4V726 A0A3B0JZA1 B4P340 A0A0A1XQ30 A0A1S3JSC1 Q9NDR2 B4Q4R0 B4IEI7 Q9V3H0 Q29KJ1 B4JZC1
A0A212EP59 L7X1G4 A0A0L7LGR6 A0A0M9A9K2 V9IAB8 A0A310SHW6 W5JGP2 E0VAJ8 A0A1B6D4X6 A0A1B6CDC6 A0A182NBH5 A0A154PA95 A0A2J7PXZ6 A0A195FWH6 A0A182J889 A0A1B6EA14 A0A182QKM7 A0A182M0H1 A0A182VTZ2 A0A158NN72 A0A2A3EK54 A0A026VZ81 A0A0C9QV11 A0A084VH37 A0A3L8DG10 A0A182R3W6 A0A067QYQ3 F4WNU5 K7J7X5 A0A182K9P8 A0A182F0Q3 A0A182YFV0 A0A2P8ZLG4 A0A182PM90 A0A1B6FSK2 A0A146LJ53 Q7Q1Q3 A0A1B0ETL7 A0A182U7Z1 A0A146LWA4 A0A182XN40 A0A182HI82 N6SU96 U4U402 A0A232FLH6 A0A0T6AZL6 D6WW09 B0WHX2 A0A088ARL5 U5EPW9 Q17BG6 A0A1S4F990 A0A1B6KTR6 A0A182GGF6 A0A023F0S3 A0A224XFN9 A0A0P4VIT5 A0A1Y1KMX5 A0A069DSR6 A0A0V0GEA2 A0A2S2QP19 A0A1Y1KI19 J9KG38 A0A336MRH6 A0A182VLF9 A0A182LM80 A0A3M6UJD4 A0A210QD94 A0A0K8UQ57 A0A034WHR1 K1P4N7 A0A195CRW1 A0A2B4RSP0 A0A0B7BH20 B3MV75 B3NAZ4 B4M987 A0A1W4V726 A0A3B0JZA1 B4P340 A0A0A1XQ30 A0A1S3JSC1 Q9NDR2 B4Q4R0 B4IEI7 Q9V3H0 Q29KJ1 B4JZC1
Pubmed
19121390
26354079
22118469
23674305
26227816
20920257
+ More
23761445 20566863 21347285 24508170 24438588 30249741 24845553 21719571 20075255 25244985 29403074 26823975 12364791 23537049 28648823 18362917 19820115 17510324 26483478 25474469 27129103 28004739 26334808 20966253 30382153 28812685 25348373 22992520 17994087 17550304 25830018 26383154 10777599 22936249 10627165 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28652364 15632085
23761445 20566863 21347285 24508170 24438588 30249741 24845553 21719571 20075255 25244985 29403074 26823975 12364791 23537049 28648823 18362917 19820115 17510324 26483478 25474469 27129103 28004739 26334808 20966253 30382153 28812685 25348373 22992520 17994087 17550304 25830018 26383154 10777599 22936249 10627165 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28652364 15632085
EMBL
BABH01020199
ODYU01003152
SOQ41544.1
GDQN01003780
JAT87274.1
KQ459232
+ More
KPJ02635.1 KQ461155 KPJ08298.1 FP102340 CBH09286.1 AGBW02013517 OWR43285.1 KC469893 AGC92696.1 JTDY01001151 KOB74728.1 KQ435720 KOX78649.1 JR037944 JR037945 AEY58048.1 AEY58049.1 KQ768821 OAD53042.1 ADMH02001555 ETN62080.1 DS235008 EEB10404.1 GEDC01016571 JAS20727.1 GEDC01026018 JAS11280.1 KQ434856 KZC08761.1 NEVH01020852 PNF21196.1 KQ981208 KYN44781.1 GEDC01002535 JAS34763.1 AXCN02000619 AXCM01005301 ADTU01021102 KZ288222 PBC32088.1 KK107672 EZA48154.1 GBYB01007594 JAG77361.1 ATLV01013130 KE524841 KFB37281.1 QOIP01000008 RLU19344.1 KK852819 KDR15630.1 GL888239 EGI64103.1 PYGN01000023 PSN57344.1 GECZ01016776 JAS52993.1 GDHC01011687 JAQ06942.1 AAAB01008980 EAA14550.4 AJWK01011181 GDHC01006685 JAQ11944.1 APCN01005650 APGK01057068 KB741277 ENN71274.1 KB631617 ERL84705.1 NNAY01000044 OXU31606.1 LJIG01016429 KRT80627.1 KQ971361 EFA08645.1 DS231940 EDS28031.1 GANO01000035 JAB59836.1 CH477322 EAT43578.1 GEBQ01025132 JAT14845.1 JXUM01061637 JXUM01061638 KQ562161 KXJ76541.1 GBBI01003642 JAC15070.1 GFTR01005141 JAW11285.1 GDKW01001791 JAI54804.1 GEZM01083777 JAV61015.1 GBGD01002118 JAC86771.1 GECL01000461 JAP05663.1 GGMS01010274 MBY79477.1 GEZM01083778 GEZM01083776 GEZM01083775 JAV61014.1 ABLF02033238 ABLF02033247 ABLF02033251 UFQT01001034 SSX28558.1 RCHS01001413 RMX53745.1 NEDP02004099 OWF46705.1 GDHF01023603 GDHF01013818 JAI28711.1 JAI38496.1 GAKP01003831 JAC55121.1 JH817690 EKC18587.1 KQ977372 KYN03232.1 LSMT01000368 PFX19275.1 HACG01044741 CEK91606.1 CH902624 EDV33140.1 CH954177 EDV58708.1 CH940654 EDW57763.1 OUUW01000010 SPP85752.1 CM000157 EDW88282.1 GBXI01015825 GBXI01001161 JAC98466.1 JAD13131.1 AB036773 BAA97120.1 CM000361 CM002910 EDX04898.1 KMY90063.1 CH480831 EDW46014.1 AF149797 AE014134 AAF03220.2 AAF53276.1 CH379061 EAL33183.2 CH916379 EDV94043.1
KPJ02635.1 KQ461155 KPJ08298.1 FP102340 CBH09286.1 AGBW02013517 OWR43285.1 KC469893 AGC92696.1 JTDY01001151 KOB74728.1 KQ435720 KOX78649.1 JR037944 JR037945 AEY58048.1 AEY58049.1 KQ768821 OAD53042.1 ADMH02001555 ETN62080.1 DS235008 EEB10404.1 GEDC01016571 JAS20727.1 GEDC01026018 JAS11280.1 KQ434856 KZC08761.1 NEVH01020852 PNF21196.1 KQ981208 KYN44781.1 GEDC01002535 JAS34763.1 AXCN02000619 AXCM01005301 ADTU01021102 KZ288222 PBC32088.1 KK107672 EZA48154.1 GBYB01007594 JAG77361.1 ATLV01013130 KE524841 KFB37281.1 QOIP01000008 RLU19344.1 KK852819 KDR15630.1 GL888239 EGI64103.1 PYGN01000023 PSN57344.1 GECZ01016776 JAS52993.1 GDHC01011687 JAQ06942.1 AAAB01008980 EAA14550.4 AJWK01011181 GDHC01006685 JAQ11944.1 APCN01005650 APGK01057068 KB741277 ENN71274.1 KB631617 ERL84705.1 NNAY01000044 OXU31606.1 LJIG01016429 KRT80627.1 KQ971361 EFA08645.1 DS231940 EDS28031.1 GANO01000035 JAB59836.1 CH477322 EAT43578.1 GEBQ01025132 JAT14845.1 JXUM01061637 JXUM01061638 KQ562161 KXJ76541.1 GBBI01003642 JAC15070.1 GFTR01005141 JAW11285.1 GDKW01001791 JAI54804.1 GEZM01083777 JAV61015.1 GBGD01002118 JAC86771.1 GECL01000461 JAP05663.1 GGMS01010274 MBY79477.1 GEZM01083778 GEZM01083776 GEZM01083775 JAV61014.1 ABLF02033238 ABLF02033247 ABLF02033251 UFQT01001034 SSX28558.1 RCHS01001413 RMX53745.1 NEDP02004099 OWF46705.1 GDHF01023603 GDHF01013818 JAI28711.1 JAI38496.1 GAKP01003831 JAC55121.1 JH817690 EKC18587.1 KQ977372 KYN03232.1 LSMT01000368 PFX19275.1 HACG01044741 CEK91606.1 CH902624 EDV33140.1 CH954177 EDV58708.1 CH940654 EDW57763.1 OUUW01000010 SPP85752.1 CM000157 EDW88282.1 GBXI01015825 GBXI01001161 JAC98466.1 JAD13131.1 AB036773 BAA97120.1 CM000361 CM002910 EDX04898.1 KMY90063.1 CH480831 EDW46014.1 AF149797 AE014134 AAF03220.2 AAF53276.1 CH379061 EAL33183.2 CH916379 EDV94043.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000037510
UP000053105
+ More
UP000000673 UP000009046 UP000075884 UP000076502 UP000235965 UP000078541 UP000075880 UP000075886 UP000075883 UP000075920 UP000005205 UP000242457 UP000053097 UP000030765 UP000279307 UP000075900 UP000027135 UP000007755 UP000002358 UP000075881 UP000069272 UP000076408 UP000245037 UP000075885 UP000007062 UP000092461 UP000075902 UP000076407 UP000075840 UP000019118 UP000030742 UP000215335 UP000007266 UP000002320 UP000005203 UP000008820 UP000069940 UP000249989 UP000007819 UP000075903 UP000075882 UP000275408 UP000242188 UP000005408 UP000078542 UP000225706 UP000007801 UP000008711 UP000008792 UP000192221 UP000268350 UP000002282 UP000085678 UP000000304 UP000001292 UP000000803 UP000001819 UP000001070
UP000000673 UP000009046 UP000075884 UP000076502 UP000235965 UP000078541 UP000075880 UP000075886 UP000075883 UP000075920 UP000005205 UP000242457 UP000053097 UP000030765 UP000279307 UP000075900 UP000027135 UP000007755 UP000002358 UP000075881 UP000069272 UP000076408 UP000245037 UP000075885 UP000007062 UP000092461 UP000075902 UP000076407 UP000075840 UP000019118 UP000030742 UP000215335 UP000007266 UP000002320 UP000005203 UP000008820 UP000069940 UP000249989 UP000007819 UP000075903 UP000075882 UP000275408 UP000242188 UP000005408 UP000078542 UP000225706 UP000007801 UP000008711 UP000008792 UP000192221 UP000268350 UP000002282 UP000085678 UP000000304 UP000001292 UP000000803 UP000001819 UP000001070
ProteinModelPortal
H9J7Z7
A0A2H1VMR1
A0A1E1WJU4
A0A194QCC5
A0A194QS01
D0AB91
+ More
A0A212EP59 L7X1G4 A0A0L7LGR6 A0A0M9A9K2 V9IAB8 A0A310SHW6 W5JGP2 E0VAJ8 A0A1B6D4X6 A0A1B6CDC6 A0A182NBH5 A0A154PA95 A0A2J7PXZ6 A0A195FWH6 A0A182J889 A0A1B6EA14 A0A182QKM7 A0A182M0H1 A0A182VTZ2 A0A158NN72 A0A2A3EK54 A0A026VZ81 A0A0C9QV11 A0A084VH37 A0A3L8DG10 A0A182R3W6 A0A067QYQ3 F4WNU5 K7J7X5 A0A182K9P8 A0A182F0Q3 A0A182YFV0 A0A2P8ZLG4 A0A182PM90 A0A1B6FSK2 A0A146LJ53 Q7Q1Q3 A0A1B0ETL7 A0A182U7Z1 A0A146LWA4 A0A182XN40 A0A182HI82 N6SU96 U4U402 A0A232FLH6 A0A0T6AZL6 D6WW09 B0WHX2 A0A088ARL5 U5EPW9 Q17BG6 A0A1S4F990 A0A1B6KTR6 A0A182GGF6 A0A023F0S3 A0A224XFN9 A0A0P4VIT5 A0A1Y1KMX5 A0A069DSR6 A0A0V0GEA2 A0A2S2QP19 A0A1Y1KI19 J9KG38 A0A336MRH6 A0A182VLF9 A0A182LM80 A0A3M6UJD4 A0A210QD94 A0A0K8UQ57 A0A034WHR1 K1P4N7 A0A195CRW1 A0A2B4RSP0 A0A0B7BH20 B3MV75 B3NAZ4 B4M987 A0A1W4V726 A0A3B0JZA1 B4P340 A0A0A1XQ30 A0A1S3JSC1 Q9NDR2 B4Q4R0 B4IEI7 Q9V3H0 Q29KJ1 B4JZC1
A0A212EP59 L7X1G4 A0A0L7LGR6 A0A0M9A9K2 V9IAB8 A0A310SHW6 W5JGP2 E0VAJ8 A0A1B6D4X6 A0A1B6CDC6 A0A182NBH5 A0A154PA95 A0A2J7PXZ6 A0A195FWH6 A0A182J889 A0A1B6EA14 A0A182QKM7 A0A182M0H1 A0A182VTZ2 A0A158NN72 A0A2A3EK54 A0A026VZ81 A0A0C9QV11 A0A084VH37 A0A3L8DG10 A0A182R3W6 A0A067QYQ3 F4WNU5 K7J7X5 A0A182K9P8 A0A182F0Q3 A0A182YFV0 A0A2P8ZLG4 A0A182PM90 A0A1B6FSK2 A0A146LJ53 Q7Q1Q3 A0A1B0ETL7 A0A182U7Z1 A0A146LWA4 A0A182XN40 A0A182HI82 N6SU96 U4U402 A0A232FLH6 A0A0T6AZL6 D6WW09 B0WHX2 A0A088ARL5 U5EPW9 Q17BG6 A0A1S4F990 A0A1B6KTR6 A0A182GGF6 A0A023F0S3 A0A224XFN9 A0A0P4VIT5 A0A1Y1KMX5 A0A069DSR6 A0A0V0GEA2 A0A2S2QP19 A0A1Y1KI19 J9KG38 A0A336MRH6 A0A182VLF9 A0A182LM80 A0A3M6UJD4 A0A210QD94 A0A0K8UQ57 A0A034WHR1 K1P4N7 A0A195CRW1 A0A2B4RSP0 A0A0B7BH20 B3MV75 B3NAZ4 B4M987 A0A1W4V726 A0A3B0JZA1 B4P340 A0A0A1XQ30 A0A1S3JSC1 Q9NDR2 B4Q4R0 B4IEI7 Q9V3H0 Q29KJ1 B4JZC1
PDB
5XPC
E-value=8.94848e-11,
Score=155
Ontologies
GO
PANTHER
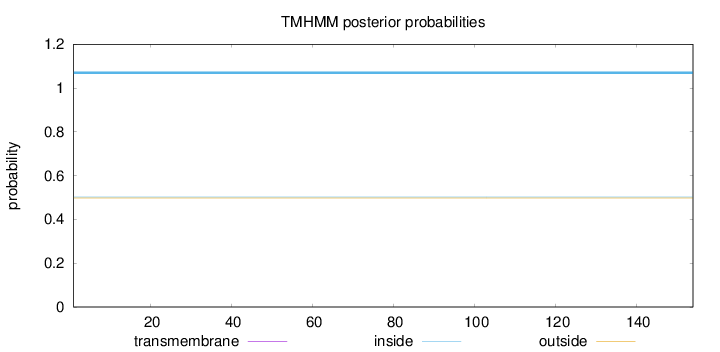
Topology
Length:
154
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0024
Exp number, first 60 AAs:
0.00012
Total prob of N-in:
0.50196
inside
1 - 154
Population Genetic Test Statistics
Pi
371.261725
Theta
150.598483
Tajima's D
4.651286
CLR
44.477326
CSRT
0.99990000499975
Interpretation
Uncertain
