Gene
KWMTBOMO10139
Pre Gene Modal
BGIBMGA005640
Annotation
Cockayne_syndrome_protein_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 3.839
Sequence
CDS
ATGGTGGACATAAACAAATATTTAGAAATGCAACTTGAATTAGCGAAAAAGCAAATTGAGAGGAAAAATAGTCAGAAAGAAGATGTATCAACTAAAAGCAAATCTGCTGCTGATTACATATCGCAAAAACAGGCAAAGACTCTGTCAAAATTTCCTCACAATCAAAAAGTAGATGTAAAAACTACTGTGACATCAGGGACTTCAACTGAAGGACACCTCGATGACATCGATTTTGTACCTTCCGATGAGGAAGACGAAAGTATCCATAATTTTCCGCTTACTGCGTGTTCTTCTCGTACCAAACAGAATATAAACCGAATTGTTGACGACGGTGATATTAAACAATACAATTTAAGGGTAGAGGTTTGGCGTGAAGGATTAAATACTGTGAAAAGCGGTACATATGAAGGATGCTCGTTAACCGCTGACATACAGTACATCCTGGATGGTAATAAAGATAGCGAAGAAATGCATGAACTCCAGAATGGTCTATTAGTACCAAAATATATTTGGAAACAGTTATATAAATTTCAAAGAACTGGTGTTAAATGGCTTTGGGAGTTGCATCAAGTGGAGTCTGGTGGATTGTTAGGTGATGAAATGGGACTTGGTAAAACAATACAAGTCATTGCATTCCTCACCGCCCTTTCAAAAACCAATTTAGGATCATGGGGCGGTCTTGGTCCGAGTATCATTATCACACCTGCAACGGTTATATATCAATGGGTATCTCATTTTCATTATTGGTGCCCCCTGCTCAGAGTAGCTGTACTGCATCACACTGGCTCTCATGCTGGTAATCACCATAAACTATTAAGGGATATACACAGTGCTCATGGAATCTTGTTAGTAACATATGCAGGCATAGTAAAGTATCATAAAGAACTCAATGCCTATAGTTGGCATTATTTAATTTTGGATGAAGGACACAAAATACGTAATCCTGATACACAAGTAAGTAAAATTGTAAAAAAAATCCAGACACCACACAGATTGTTAATAACAGGTTCACCAATGCAGAACAGTTTGCAGGAACTGTGGTCATTATTTGATTTTATGAGGCCAGGTTTGTTGGGGACATACAATGCATTCATAGAACATTTTGCAGTTCCAATCACTCAAGGAGGCTATGCTAATGCTACTGAAATACAGGAAGCTACTGCCATGGAAATAGCTATTGCTTTGAAAAATATGATCACTCCTTACATGCTGAGGAGAACTAAGGCTGAGGTACAAGGACATATCAAACTCCCAGAAAAGAATGAACAAGTTCTATTCTGTTCTTTAACACAAGAGCAAAGAGATCTTTATATGGGTTACCTTATGAGTAGTACAGTGAGAAGCATCTTAGACAAAGAAAATAAATTTGGAAATCCATTAAGAGCAAGGCTACTAGTGGCTCTGTCAACGCTAAGGAAAATATGCAATCATCCTGATATTTATCTCTATGAAGCTCAAGAAGATGAAGAACAGATAGATGAAGAGATATTTGGCCATTGGAAGAGATCTGGAAAAATGACTGTTGTTAATTCATTACTTAAAATATGGAAAAAACAGGGACATAGAGCTCTCATATTCTCTCAGTCAAGAGTAATGCTGTGTCTGTTAGAACAGTATTTACAAAATCAAGGCTTTAAATACCTCAAAATGGATGGTACAGTTAATGTTGGCCAAAGGCAAAACTTGATTAAAACTTTTAATGATAACACTGATTATTTAGTTTTTCTTTCAACAACTAGAGTAGGTGGCTTAGGTGTAAATTTAACTGGAGCCGACAGAGTGATTATATATGATCCTGATTGGAATCCTGCAACAGATGATCAAGCAAAGGAAAGAGCATGGAGGATAGGCCAATCTAGAAATGTAACAGTTTATAGATTGTTGTCAGCTGGGACTATAGAGGAAAAGATTTATCAGAGGCAGATATTCAAACATTTCTTGAGTAATAAAATTTTTGTGGACCCAAATCAAAAGAATTTTTTGACCACCAGTACATTACAGGGACTGTTCACTCTTGAAGAGCTTAACCGTGACGGAGATACTGAAACTGCATCAATTTTTAAACACACCAAAATTATCAATAAAACCAATAAGGATGATAATGACTTGGATATTTCATCTAAAACAAAGTTAGAAGCCATGAAAAGATTAGCAAAAGAAATAAGTAAAAAAATATCAAAAGAATCCATGAAGGGAACACAGGAAAGTGATCCACGTGAAGAATATAAAAAGAAAAGAGAACAATTATTATGTCCACCTAAAGTGGAAGAACCAGATATTAATTTAGTTGATGATAAAATAACCAATGTAGCTTTTGAAGATGTTTTGTCAGAATTAGATATTGTAAAAATGCATGCAAAGAAAGATTATGAAGAACAATTGTTGAAAAGTGTACTAGAAGATAGTGAACAAGATATTAAAGAAGTAGATGAAGAGAAACCTACCAACATTGACAACTCTCAACTTGAAACTAATGAAAGTACTGTAAAAGATATTAAACAAAGGCATAAAAAATCACATAAAATAAAGCGCAAAAGAAGATTATCATCTTCCATTGATGATGGACTCAAAAATTTAGTAACTGTAAAGAAAGTAAAGAAAAGGATTATAGTAAAAATGAAGTAA
Protein
MVDINKYLEMQLELAKKQIERKNSQKEDVSTKSKSAADYISQKQAKTLSKFPHNQKVDVKTTVTSGTSTEGHLDDIDFVPSDEEDESIHNFPLTACSSRTKQNINRIVDDGDIKQYNLRVEVWREGLNTVKSGTYEGCSLTADIQYILDGNKDSEEMHELQNGLLVPKYIWKQLYKFQRTGVKWLWELHQVESGGLLGDEMGLGKTIQVIAFLTALSKTNLGSWGGLGPSIIITPATVIYQWVSHFHYWCPLLRVAVLHHTGSHAGNHHKLLRDIHSAHGILLVTYAGIVKYHKELNAYSWHYLILDEGHKIRNPDTQVSKIVKKIQTPHRLLITGSPMQNSLQELWSLFDFMRPGLLGTYNAFIEHFAVPITQGGYANATEIQEATAMEIAIALKNMITPYMLRRTKAEVQGHIKLPEKNEQVLFCSLTQEQRDLYMGYLMSSTVRSILDKENKFGNPLRARLLVALSTLRKICNHPDIYLYEAQEDEEQIDEEIFGHWKRSGKMTVVNSLLKIWKKQGHRALIFSQSRVMLCLLEQYLQNQGFKYLKMDGTVNVGQRQNLIKTFNDNTDYLVFLSTTRVGGLGVNLTGADRVIIYDPDWNPATDDQAKERAWRIGQSRNVTVYRLLSAGTIEEKIYQRQIFKHFLSNKIFVDPNQKNFLTTSTLQGLFTLEELNRDGDTETASIFKHTKIINKTNKDDNDLDISSKTKLEAMKRLAKEISKKISKESMKGTQESDPREEYKKKREQLLCPPKVEEPDINLVDDKITNVAFEDVLSELDIVKMHAKKDYEEQLLKSVLEDSEQDIKEVDEEKPTNIDNSQLETNESTVKDIKQRHKKSHKIKRKRRLSSSIDDGLKNLVTVKKVKKRIIVKMK
Summary
Uniprot
H9J7Z8
A0A0L7LLZ6
A0A2A4JD02
E3UKJ6
L7X1P1
A0A194QXK5
+ More
A0A194QAY5 D0AB88 A0A2H1VL62 A0A212EP84 D6WQR8 A0A154PN39 A0A088AQ00 K7J3J4 A0A232FE21 A0A2A3EJZ4 A0A0L7QLV0 A0A1Y1KCK3 A0A2J7Q3N2 A0A0C9RBF4 A0A151WWB2 A0A1Y1KDE8 A0A026W8Z2 A0A1B6DRP9 F4WJ78 A0A067QXI6 A0A158NED4 A0A195BX22 A0A195CAG4 A0A151J6K1 A0A2Z5U5Z9 H0ZD23 A0A218UXI1 U3JYN5 A0A091N177 A0A091FPW4 A0A091ECV7 A0A093IEP1 A0A091KR04 A0A093QHQ1 E1BYA8 A0A091JDH4 A0A091LPB5 A0A3L8SKU3 A0A091UMZ7 A0A1V4KKV1 A0A0A0AQU7 F1R294 G1N7W0 A0A091Q912 A0A091PS46 A0A091UHQ3 A0A093R180 A0A091TAS4 A0A093HJK3 A0A094KYG1 U3J9Q7 A0A091IEY6 A0A0P6GYJ3 A0A0P5ZPG8 A0A0P5DCD7 A0A0P6AVW5 A0A165A3J8 A0A3B4B6R9 A0A0M8ZQ55 A0A0P5D7G3 W5M9D1 R0JQI4 A0A087QZ10 A0A0P5CLD2 A0A0P5CLI3 A0A0P5D036 A0A0N8AA90 A0A091GMZ3 A0A093NG49 A0A0P5DVE3 L5KAG3 A0A3Q2Z7L0 A0A3B4BMT4 A0A1L8FKT9 A0A1W4XWV5 A0A3Q2FG36 A0A1W4XVY9
A0A194QAY5 D0AB88 A0A2H1VL62 A0A212EP84 D6WQR8 A0A154PN39 A0A088AQ00 K7J3J4 A0A232FE21 A0A2A3EJZ4 A0A0L7QLV0 A0A1Y1KCK3 A0A2J7Q3N2 A0A0C9RBF4 A0A151WWB2 A0A1Y1KDE8 A0A026W8Z2 A0A1B6DRP9 F4WJ78 A0A067QXI6 A0A158NED4 A0A195BX22 A0A195CAG4 A0A151J6K1 A0A2Z5U5Z9 H0ZD23 A0A218UXI1 U3JYN5 A0A091N177 A0A091FPW4 A0A091ECV7 A0A093IEP1 A0A091KR04 A0A093QHQ1 E1BYA8 A0A091JDH4 A0A091LPB5 A0A3L8SKU3 A0A091UMZ7 A0A1V4KKV1 A0A0A0AQU7 F1R294 G1N7W0 A0A091Q912 A0A091PS46 A0A091UHQ3 A0A093R180 A0A091TAS4 A0A093HJK3 A0A094KYG1 U3J9Q7 A0A091IEY6 A0A0P6GYJ3 A0A0P5ZPG8 A0A0P5DCD7 A0A0P6AVW5 A0A165A3J8 A0A3B4B6R9 A0A0M8ZQ55 A0A0P5D7G3 W5M9D1 R0JQI4 A0A087QZ10 A0A0P5CLD2 A0A0P5CLI3 A0A0P5D036 A0A0N8AA90 A0A091GMZ3 A0A093NG49 A0A0P5DVE3 L5KAG3 A0A3Q2Z7L0 A0A3B4BMT4 A0A1L8FKT9 A0A1W4XWV5 A0A3Q2FG36 A0A1W4XVY9
Pubmed
EMBL
BABH01020201
JTDY01000686
KOB76216.1
NWSH01001832
PCG69965.1
HM449870
+ More
ADO33005.1 KC469893 AGC92693.1 KQ461155 KPJ08306.1 KQ459232 KPJ02627.1 FP102340 CBH09283.1 ODYU01003152 SOQ41541.1 AGBW02013517 OWR43289.1 KQ971354 EFA07002.2 KQ434998 KZC13269.1 NNAY01000390 OXU28718.1 KZ288232 PBC31516.1 KQ414905 KOC59628.1 GEZM01088381 JAV58228.1 NEVH01019065 PNF23187.1 GBYB01013814 JAG83581.1 KQ982696 KYQ51971.1 GEZM01088380 JAV58230.1 KK107388 EZA51499.1 GEDC01008947 JAS28351.1 GL888182 EGI65656.1 KK853134 KDR10852.1 ADTU01013257 ADTU01013258 KQ976401 KYM92518.1 KQ978023 KYM97864.1 KQ979851 KYN18853.1 FX985816 BBA93703.1 ABQF01027374 ABQF01027375 ABQF01027376 ABQF01027377 ABQF01027378 ABQF01027379 ABQF01027380 MUZQ01000106 OWK58211.1 AGTO01000148 KK839952 KFP82644.1 KL447250 KFO71011.1 KK718105 KFO54269.1 KL215552 KFV65211.1 KK755005 KFP43029.1 KL672532 KFW85930.1 AADN05000339 KK501886 KFP18652.1 KK505808 KFP61313.1 QUSF01000014 RLW03973.1 KL409688 KFQ91270.1 LSYS01003057 OPJ84447.1 KL872592 KGL96282.1 CR762493 KK690606 KFQ16455.1 KK645687 KFP98775.1 KK436209 KFQ90314.1 KL424518 KFW89922.1 KK443449 KFQ70947.1 KL206495 KFV82828.1 KL267025 KFZ62302.1 ADON01089224 ADON01089225 KL218641 KFP07189.1 GDIQ01039149 JAN55588.1 GDIP01041139 JAM62576.1 GDIP01159619 JAJ63783.1 GDIP01025796 JAM77919.1 LRGB01000642 KZS17136.1 KQ435922 KOX68657.1 GDIP01160127 JAJ63275.1 AHAT01025219 AHAT01025220 KB743335 EOA99371.1 KL226002 KFM06464.1 GDIP01168821 JAJ54581.1 GDIP01168822 JAJ54580.1 GDIP01168116 GDIP01167218 JAJ56184.1 GDIP01167217 JAJ56185.1 KL504946 KFO84504.1 KL224536 KFW60990.1 GDIP01168115 JAJ55287.1 KB030894 ELK08347.1 CM004478 OCT72187.1
ADO33005.1 KC469893 AGC92693.1 KQ461155 KPJ08306.1 KQ459232 KPJ02627.1 FP102340 CBH09283.1 ODYU01003152 SOQ41541.1 AGBW02013517 OWR43289.1 KQ971354 EFA07002.2 KQ434998 KZC13269.1 NNAY01000390 OXU28718.1 KZ288232 PBC31516.1 KQ414905 KOC59628.1 GEZM01088381 JAV58228.1 NEVH01019065 PNF23187.1 GBYB01013814 JAG83581.1 KQ982696 KYQ51971.1 GEZM01088380 JAV58230.1 KK107388 EZA51499.1 GEDC01008947 JAS28351.1 GL888182 EGI65656.1 KK853134 KDR10852.1 ADTU01013257 ADTU01013258 KQ976401 KYM92518.1 KQ978023 KYM97864.1 KQ979851 KYN18853.1 FX985816 BBA93703.1 ABQF01027374 ABQF01027375 ABQF01027376 ABQF01027377 ABQF01027378 ABQF01027379 ABQF01027380 MUZQ01000106 OWK58211.1 AGTO01000148 KK839952 KFP82644.1 KL447250 KFO71011.1 KK718105 KFO54269.1 KL215552 KFV65211.1 KK755005 KFP43029.1 KL672532 KFW85930.1 AADN05000339 KK501886 KFP18652.1 KK505808 KFP61313.1 QUSF01000014 RLW03973.1 KL409688 KFQ91270.1 LSYS01003057 OPJ84447.1 KL872592 KGL96282.1 CR762493 KK690606 KFQ16455.1 KK645687 KFP98775.1 KK436209 KFQ90314.1 KL424518 KFW89922.1 KK443449 KFQ70947.1 KL206495 KFV82828.1 KL267025 KFZ62302.1 ADON01089224 ADON01089225 KL218641 KFP07189.1 GDIQ01039149 JAN55588.1 GDIP01041139 JAM62576.1 GDIP01159619 JAJ63783.1 GDIP01025796 JAM77919.1 LRGB01000642 KZS17136.1 KQ435922 KOX68657.1 GDIP01160127 JAJ63275.1 AHAT01025219 AHAT01025220 KB743335 EOA99371.1 KL226002 KFM06464.1 GDIP01168821 JAJ54581.1 GDIP01168822 JAJ54580.1 GDIP01168116 GDIP01167218 JAJ56184.1 GDIP01167217 JAJ56185.1 KL504946 KFO84504.1 KL224536 KFW60990.1 GDIP01168115 JAJ55287.1 KB030894 ELK08347.1 CM004478 OCT72187.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000007266 UP000076502 UP000005203 UP000002358 UP000215335 UP000242457 UP000053825 UP000235965 UP000075809 UP000053097 UP000007755 UP000027135 UP000005205 UP000078540 UP000078542 UP000078492 UP000007754 UP000197619 UP000016665 UP000053760 UP000052976 UP000053875 UP000053258 UP000000539 UP000053119 UP000276834 UP000053283 UP000190648 UP000053858 UP000000437 UP000001645 UP000053584 UP000016666 UP000054308 UP000076858 UP000261520 UP000053105 UP000018468 UP000053286 UP000054081 UP000010552 UP000264820 UP000261440 UP000186698 UP000192224 UP000265020
UP000007266 UP000076502 UP000005203 UP000002358 UP000215335 UP000242457 UP000053825 UP000235965 UP000075809 UP000053097 UP000007755 UP000027135 UP000005205 UP000078540 UP000078542 UP000078492 UP000007754 UP000197619 UP000016665 UP000053760 UP000052976 UP000053875 UP000053258 UP000000539 UP000053119 UP000276834 UP000053283 UP000190648 UP000053858 UP000000437 UP000001645 UP000053584 UP000016666 UP000054308 UP000076858 UP000261520 UP000053105 UP000018468 UP000053286 UP000054081 UP000010552 UP000264820 UP000261440 UP000186698 UP000192224 UP000265020
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
CDD
ProteinModelPortal
H9J7Z8
A0A0L7LLZ6
A0A2A4JD02
E3UKJ6
L7X1P1
A0A194QXK5
+ More
A0A194QAY5 D0AB88 A0A2H1VL62 A0A212EP84 D6WQR8 A0A154PN39 A0A088AQ00 K7J3J4 A0A232FE21 A0A2A3EJZ4 A0A0L7QLV0 A0A1Y1KCK3 A0A2J7Q3N2 A0A0C9RBF4 A0A151WWB2 A0A1Y1KDE8 A0A026W8Z2 A0A1B6DRP9 F4WJ78 A0A067QXI6 A0A158NED4 A0A195BX22 A0A195CAG4 A0A151J6K1 A0A2Z5U5Z9 H0ZD23 A0A218UXI1 U3JYN5 A0A091N177 A0A091FPW4 A0A091ECV7 A0A093IEP1 A0A091KR04 A0A093QHQ1 E1BYA8 A0A091JDH4 A0A091LPB5 A0A3L8SKU3 A0A091UMZ7 A0A1V4KKV1 A0A0A0AQU7 F1R294 G1N7W0 A0A091Q912 A0A091PS46 A0A091UHQ3 A0A093R180 A0A091TAS4 A0A093HJK3 A0A094KYG1 U3J9Q7 A0A091IEY6 A0A0P6GYJ3 A0A0P5ZPG8 A0A0P5DCD7 A0A0P6AVW5 A0A165A3J8 A0A3B4B6R9 A0A0M8ZQ55 A0A0P5D7G3 W5M9D1 R0JQI4 A0A087QZ10 A0A0P5CLD2 A0A0P5CLI3 A0A0P5D036 A0A0N8AA90 A0A091GMZ3 A0A093NG49 A0A0P5DVE3 L5KAG3 A0A3Q2Z7L0 A0A3B4BMT4 A0A1L8FKT9 A0A1W4XWV5 A0A3Q2FG36 A0A1W4XVY9
A0A194QAY5 D0AB88 A0A2H1VL62 A0A212EP84 D6WQR8 A0A154PN39 A0A088AQ00 K7J3J4 A0A232FE21 A0A2A3EJZ4 A0A0L7QLV0 A0A1Y1KCK3 A0A2J7Q3N2 A0A0C9RBF4 A0A151WWB2 A0A1Y1KDE8 A0A026W8Z2 A0A1B6DRP9 F4WJ78 A0A067QXI6 A0A158NED4 A0A195BX22 A0A195CAG4 A0A151J6K1 A0A2Z5U5Z9 H0ZD23 A0A218UXI1 U3JYN5 A0A091N177 A0A091FPW4 A0A091ECV7 A0A093IEP1 A0A091KR04 A0A093QHQ1 E1BYA8 A0A091JDH4 A0A091LPB5 A0A3L8SKU3 A0A091UMZ7 A0A1V4KKV1 A0A0A0AQU7 F1R294 G1N7W0 A0A091Q912 A0A091PS46 A0A091UHQ3 A0A093R180 A0A091TAS4 A0A093HJK3 A0A094KYG1 U3J9Q7 A0A091IEY6 A0A0P6GYJ3 A0A0P5ZPG8 A0A0P5DCD7 A0A0P6AVW5 A0A165A3J8 A0A3B4B6R9 A0A0M8ZQ55 A0A0P5D7G3 W5M9D1 R0JQI4 A0A087QZ10 A0A0P5CLD2 A0A0P5CLI3 A0A0P5D036 A0A0N8AA90 A0A091GMZ3 A0A093NG49 A0A0P5DVE3 L5KAG3 A0A3Q2Z7L0 A0A3B4BMT4 A0A1L8FKT9 A0A1W4XWV5 A0A3Q2FG36 A0A1W4XVY9
PDB
5VVR
E-value=2.78165e-132,
Score=1212
Ontologies
GO
GO:0005524
GO:0045739
GO:0035264
GO:0032786
GO:0000303
GO:0010224
GO:0003682
GO:0003677
GO:0005730
GO:0006283
GO:0006290
GO:0008022
GO:0007257
GO:0044877
GO:0008630
GO:0010165
GO:0006284
GO:0010332
GO:0006362
GO:0008023
GO:0008094
GO:0007256
GO:0030296
GO:0045494
GO:0009636
GO:0047485
GO:0006974
GO:0006468
GO:0004535
GO:0003723
GO:0005634
GO:0005737
GO:0006402
GO:0046872
GO:0003824
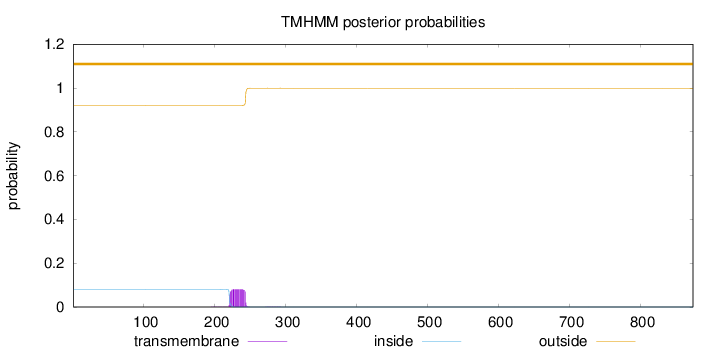
Topology
Length:
874
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.85405
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08063
outside
1 - 874
Population Genetic Test Statistics
Pi
19.739753
Theta
33.3508
Tajima's D
-1.157369
CLR
111.178279
CSRT
0.112094395280236
Interpretation
Uncertain
