Gene
KWMTBOMO10138 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005641
Annotation
putative_mitogen-activated_protein_kinase_(MAPKK)_[Heliconius_melpomene]
Full name
Diacylglycerol kinase
Location in the cell
Nuclear Reliability : 2.758
Sequence
CDS
ATGTCGAGAAGAAAACTGCCGCCCCCGGGCTTTAACTTTAAGACTCAAACGGAACAAGTTGTGACTCCACCAGGGAATTTAGATAAACAAACAACAATTACAGTTGATGACCGAACATTCACTGTACATGCTGATGATTTGGTAAAGATTTGTGATCTTGGCCGAGGAGCATATGGCATTGTTGAGAAGATGCGCCACCTACCTAGCAACACTATTATGGCTGTTAAGAGAATCACAGCCACATTCAACAATCAATCTTTGGAACAAAAACGTTTGTTAATGGATCTTGATGTATCAATGAGAGCAAGGAAGGGATGTTTGGATTTGTATGGAAGTAATGGATATGAGTTTAGACAAATTTTACACAAAAGTCTACAAAATGATCGTACCATACCTGAGAATATTCTTGGAAAGATAGCATTTTCGGTTGTTAGTGCTTTACATTATTTATACTCCAAACTCAGAGTCATACATAGAGATGTGAAACCATCAAATATATTGATCAACAGAAAGGGTGAAGTGAAGATGTGTGACTTTGGCATTTCTGGTCATCTGGTAGATTCTGTTGCAAAAACAATTGATGCTGGCTGTAAACCTTATATGGCACCAGAACGAATTGATCCAACTGGAAATCCTGGGCAATATGACATTCGAAGTGACGTGTGGTCTCTGGGAATATCCATGATTGAATTAGCAACAGGAACTTTCCCATACAACACATGGGGCACACCTTTCGAGCAACTAAAACAAGTTGTTAAAGATGATCCACCAAGGCTTCCAGTGGGAAAATTTTCAAACGAGTTTGAGGATTTAATAGTGCAATGCTTACAGAAAGACTATAGACTCAGACCAAATTACGATGCATTGTTAGTGCATCCATTCTGCCAGGAACACAGTCAAAAGGAAACCGATGTTGCATCTTTTGTTCAGGAAATATTAGATTTAGCAGGCGATGCTTAG
Protein
MSRRKLPPPGFNFKTQTEQVVTPPGNLDKQTTITVDDRTFTVHADDLVKICDLGRGAYGIVEKMRHLPSNTIMAVKRITATFNNQSLEQKRLLMDLDVSMRARKGCLDLYGSNGYEFRQILHKSLQNDRTIPENILGKIAFSVVSALHYLYSKLRVIHRDVKPSNILINRKGEVKMCDFGISGHLVDSVAKTIDAGCKPYMAPERIDPTGNPGQYDIRSDVWSLGISMIELATGTFPYNTWGTPFEQLKQVVKDDPPRLPVGKFSNEFEDLIVQCLQKDYRLRPNYDALLVHPFCQEHSQKETDVASFVQEILDLAGDA
Summary
Catalytic Activity
a 1,2-diacyl-sn-glycerol + ATP = a 1,2-diacyl-sn-glycero-3-phosphate + ADP + H(+)
Similarity
Belongs to the protein kinase superfamily.
Belongs to the eukaryotic diacylglycerol kinase family.
Belongs to the eukaryotic diacylglycerol kinase family.
Uniprot
H9J7Z9
A0A2A4JEF0
D0AB87
L7WW91
A0A2H1VVK4
A0A212EP69
+ More
A0A194QAR5 A0A194QTH5 S4NSS2 A0A0L7LPN6 A0A1Y1JW72 A0A2P8Y5R8 A0A1W4XB76 A0A1B6IB42 A0A1B6GET5 A0A067QN57 A0A1B6L3U1 A9XK84 A0A0T6AYY0 A0A0M9AAQ5 E0VQ74 A0A026WS53 A0A158NNP5 A0A023ER47 E2B907 A0A151HZL0 A0A1L8DM37 E2AZT9 A0A2J7PXF6 Q17KN4 Q67EX4 U5EW25 A0A069DYN1 A0A1L8DMC4 A0A0J7KV34 A0A0K8TSJ2 A0A0C9QM81 A0A224XTF1 A0A0P4VRJ8 R4G3V7 A0A1I8PC03 A0A1I8M2L4 T1PDH9 A0A151X7T9 A0A1Q3F5W1 A0A154P2K1 A0A2A3E1J1 A0A0V0G9F5 A0A151J3E7 A0A0L7QK85 B0W7R1 V5GF02 J9JYM3 A0A336LUS6 D6WX74 K7J2E0 A0A232F0F6 A0A1A9VNI1 A0A1B0AFB7 D3TLE9 A0A1A9WVR0 A0A088AVZ4 A0A0K8UXF9 A0A034WHR3 A0A2H8TDS5 A0A195FS05 A0A0A1XEX4 A0A310SX15 A0A2R7W8R2 B4JLY8 A0A1J1IV25 A0A0L0BUV4 T1IUF7 B4L621 W8B7L0 B4Q205 B3MS42 B4GW95 A0A1W4VK79 A0A3B0KRW6 B4M728 Q29IB7 B3NX72 A0A0M4F9A4 B4NUD3 B4IGM0 O62602 E9H2U1 Q9U983 B4NEN3 A0A0N8CT73 A0A0N8DCF9 A0A0P5L8K9 A0A0N7ZKQ6 A0A0N8AGI5 A0A0P5Y8M6 U4U2E7 A0A0N8DX38 N6UGG3 A0A0P5EKH4
A0A194QAR5 A0A194QTH5 S4NSS2 A0A0L7LPN6 A0A1Y1JW72 A0A2P8Y5R8 A0A1W4XB76 A0A1B6IB42 A0A1B6GET5 A0A067QN57 A0A1B6L3U1 A9XK84 A0A0T6AYY0 A0A0M9AAQ5 E0VQ74 A0A026WS53 A0A158NNP5 A0A023ER47 E2B907 A0A151HZL0 A0A1L8DM37 E2AZT9 A0A2J7PXF6 Q17KN4 Q67EX4 U5EW25 A0A069DYN1 A0A1L8DMC4 A0A0J7KV34 A0A0K8TSJ2 A0A0C9QM81 A0A224XTF1 A0A0P4VRJ8 R4G3V7 A0A1I8PC03 A0A1I8M2L4 T1PDH9 A0A151X7T9 A0A1Q3F5W1 A0A154P2K1 A0A2A3E1J1 A0A0V0G9F5 A0A151J3E7 A0A0L7QK85 B0W7R1 V5GF02 J9JYM3 A0A336LUS6 D6WX74 K7J2E0 A0A232F0F6 A0A1A9VNI1 A0A1B0AFB7 D3TLE9 A0A1A9WVR0 A0A088AVZ4 A0A0K8UXF9 A0A034WHR3 A0A2H8TDS5 A0A195FS05 A0A0A1XEX4 A0A310SX15 A0A2R7W8R2 B4JLY8 A0A1J1IV25 A0A0L0BUV4 T1IUF7 B4L621 W8B7L0 B4Q205 B3MS42 B4GW95 A0A1W4VK79 A0A3B0KRW6 B4M728 Q29IB7 B3NX72 A0A0M4F9A4 B4NUD3 B4IGM0 O62602 E9H2U1 Q9U983 B4NEN3 A0A0N8CT73 A0A0N8DCF9 A0A0P5L8K9 A0A0N7ZKQ6 A0A0N8AGI5 A0A0P5Y8M6 U4U2E7 A0A0N8DX38 N6UGG3 A0A0P5EKH4
EC Number
2.7.1.107
Pubmed
19121390
23674305
22118469
26354079
23622113
26227816
+ More
28004739 29403074 24845553 17706772 20566863 24508170 30249741 21347285 24945155 26483478 20798317 17510324 14986920 26334808 26369729 27129103 25315136 18362917 19820115 20075255 28648823 20353571 25348373 25830018 17994087 26108605 24495485 17550304 15632085 9584193 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 10364162 23537049
28004739 29403074 24845553 17706772 20566863 24508170 30249741 21347285 24945155 26483478 20798317 17510324 14986920 26334808 26369729 27129103 25315136 18362917 19820115 20075255 28648823 20353571 25348373 25830018 17994087 26108605 24495485 17550304 15632085 9584193 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21292972 10364162 23537049
EMBL
BABH01020201
NWSH01001832
PCG69964.1
FP102340
CBH09282.1
KC469893
+ More
AGC92692.1 ODYU01004701 SOQ44860.1 AGBW02013517 OWR43290.1 KQ459232 KPJ02628.1 KQ461155 KPJ08305.1 GAIX01013942 JAA78618.1 JTDY01000391 KOB77400.1 GEZM01099000 JAV53582.1 PYGN01000896 PSN39599.1 GECU01037056 GECU01036185 GECU01035622 GECU01034966 GECU01026631 GECU01023613 GECU01023593 GECU01021463 GECU01018946 GECU01017131 GECU01006000 GECU01005905 GECU01004573 GECU01003580 GECU01003023 GECU01002428 JAS70650.1 JAS71521.1 JAS72084.1 JAS72740.1 JAS81075.1 JAS84093.1 JAS84113.1 JAS86243.1 JAS88760.1 JAS90575.1 JAT01707.1 JAT01802.1 JAT03134.1 JAT04127.1 JAT04684.1 JAT05279.1 GECZ01008848 JAS60921.1 KK853134 KDR10863.1 GEBQ01021620 GEBQ01001690 JAT18357.1 JAT38287.1 EF491249 ABR28347.1 LJIG01022489 KRT80305.1 KQ435711 KOX79423.1 AAZO01004340 DS235392 EEB15530.1 KK107119 QOIP01000009 EZA58783.1 RLU18329.1 ADTU01002755 ADTU01002756 JXUM01007305 JXUM01007306 JXUM01007307 GAPW01002484 KQ560237 JAC11114.1 KXJ83678.1 GL446405 EFN87841.1 KQ976693 KYM77658.1 GFDF01006558 JAV07526.1 GL444277 EFN61068.1 NEVH01020858 PNF21021.1 CH477222 EAT47270.1 AY358075 AAQ68075.1 GANO01003151 JAB56720.1 GBGD01002100 JAC86789.1 GFDF01006559 JAV07525.1 LBMM01002940 KMQ94158.1 GDAI01000688 JAI16915.1 GBYB01001722 JAG71489.1 GFTR01005177 JAW11249.1 GDKW01001556 JAI55039.1 GAHY01001331 JAA76179.1 KA646749 AFP61378.1 KQ982431 KYQ56442.1 GFDL01012090 JAV22955.1 KQ434789 KZC05350.1 KZ288451 PBC25568.1 GECL01001404 JAP04720.1 KQ980284 KYN16962.1 KQ414989 KOC58975.1 DS231855 EDS38205.1 GALX01005912 JAB62554.1 ABLF02040695 UFQS01000212 UFQT01000212 SSX01406.1 SSX21786.1 KQ971361 EFA08029.1 AAZX01007551 NNAY01001391 OXU24141.1 CCAG010006445 EZ422251 ADD18527.1 GDHF01021048 JAI31266.1 GAKP01004758 JAC54194.1 GFXV01000415 MBW12220.1 KQ981285 KYN43211.1 GBXI01010246 GBXI01004816 JAD04046.1 JAD09476.1 KQ759840 OAD62646.1 KK854406 PTY15551.1 CH916371 EDV91749.1 CVRI01000061 CRL04061.1 JRES01001303 KNC23773.1 JH431533 CH933812 EDW05817.1 GAMC01009365 GAMC01009363 JAB97190.1 CM000162 EDX01526.1 CH902622 EDV34597.1 CH479194 EDW26979.1 OUUW01000021 SPP89409.1 CH940653 EDW62595.1 CH379063 EAL32736.2 CH954180 EDV47244.1 CP012528 ALC48757.1 CH983901 EDX16580.1 CH480836 EDW48970.1 AF035549 AF035550 AF041136 AE014298 AY052103 AAC39033.1 AAC39034.1 AAF22365.1 AAF48223.1 AAK93527.1 GL732587 EFX73950.1 AJ238572 CAB45101.1 CH964239 EDW82202.1 GDIP01101263 JAM02452.1 GDIP01047375 JAM56340.1 GDIQ01173279 JAK78446.1 GDIP01235889 JAI87512.1 GDIP01149657 JAJ73745.1 GDIP01062764 JAM40951.1 KB632013 ERL88044.1 GDIQ01083364 JAN11373.1 APGK01036393 KB740939 ENN77747.1 GDIQ01268410 JAJ83314.1
AGC92692.1 ODYU01004701 SOQ44860.1 AGBW02013517 OWR43290.1 KQ459232 KPJ02628.1 KQ461155 KPJ08305.1 GAIX01013942 JAA78618.1 JTDY01000391 KOB77400.1 GEZM01099000 JAV53582.1 PYGN01000896 PSN39599.1 GECU01037056 GECU01036185 GECU01035622 GECU01034966 GECU01026631 GECU01023613 GECU01023593 GECU01021463 GECU01018946 GECU01017131 GECU01006000 GECU01005905 GECU01004573 GECU01003580 GECU01003023 GECU01002428 JAS70650.1 JAS71521.1 JAS72084.1 JAS72740.1 JAS81075.1 JAS84093.1 JAS84113.1 JAS86243.1 JAS88760.1 JAS90575.1 JAT01707.1 JAT01802.1 JAT03134.1 JAT04127.1 JAT04684.1 JAT05279.1 GECZ01008848 JAS60921.1 KK853134 KDR10863.1 GEBQ01021620 GEBQ01001690 JAT18357.1 JAT38287.1 EF491249 ABR28347.1 LJIG01022489 KRT80305.1 KQ435711 KOX79423.1 AAZO01004340 DS235392 EEB15530.1 KK107119 QOIP01000009 EZA58783.1 RLU18329.1 ADTU01002755 ADTU01002756 JXUM01007305 JXUM01007306 JXUM01007307 GAPW01002484 KQ560237 JAC11114.1 KXJ83678.1 GL446405 EFN87841.1 KQ976693 KYM77658.1 GFDF01006558 JAV07526.1 GL444277 EFN61068.1 NEVH01020858 PNF21021.1 CH477222 EAT47270.1 AY358075 AAQ68075.1 GANO01003151 JAB56720.1 GBGD01002100 JAC86789.1 GFDF01006559 JAV07525.1 LBMM01002940 KMQ94158.1 GDAI01000688 JAI16915.1 GBYB01001722 JAG71489.1 GFTR01005177 JAW11249.1 GDKW01001556 JAI55039.1 GAHY01001331 JAA76179.1 KA646749 AFP61378.1 KQ982431 KYQ56442.1 GFDL01012090 JAV22955.1 KQ434789 KZC05350.1 KZ288451 PBC25568.1 GECL01001404 JAP04720.1 KQ980284 KYN16962.1 KQ414989 KOC58975.1 DS231855 EDS38205.1 GALX01005912 JAB62554.1 ABLF02040695 UFQS01000212 UFQT01000212 SSX01406.1 SSX21786.1 KQ971361 EFA08029.1 AAZX01007551 NNAY01001391 OXU24141.1 CCAG010006445 EZ422251 ADD18527.1 GDHF01021048 JAI31266.1 GAKP01004758 JAC54194.1 GFXV01000415 MBW12220.1 KQ981285 KYN43211.1 GBXI01010246 GBXI01004816 JAD04046.1 JAD09476.1 KQ759840 OAD62646.1 KK854406 PTY15551.1 CH916371 EDV91749.1 CVRI01000061 CRL04061.1 JRES01001303 KNC23773.1 JH431533 CH933812 EDW05817.1 GAMC01009365 GAMC01009363 JAB97190.1 CM000162 EDX01526.1 CH902622 EDV34597.1 CH479194 EDW26979.1 OUUW01000021 SPP89409.1 CH940653 EDW62595.1 CH379063 EAL32736.2 CH954180 EDV47244.1 CP012528 ALC48757.1 CH983901 EDX16580.1 CH480836 EDW48970.1 AF035549 AF035550 AF041136 AE014298 AY052103 AAC39033.1 AAC39034.1 AAF22365.1 AAF48223.1 AAK93527.1 GL732587 EFX73950.1 AJ238572 CAB45101.1 CH964239 EDW82202.1 GDIP01101263 JAM02452.1 GDIP01047375 JAM56340.1 GDIQ01173279 JAK78446.1 GDIP01235889 JAI87512.1 GDIP01149657 JAJ73745.1 GDIP01062764 JAM40951.1 KB632013 ERL88044.1 GDIQ01083364 JAN11373.1 APGK01036393 KB740939 ENN77747.1 GDIQ01268410 JAJ83314.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000245037 UP000192223 UP000027135 UP000053105 UP000009046 UP000053097 UP000279307 UP000005205 UP000069940 UP000249989 UP000008237 UP000078540 UP000000311 UP000235965 UP000008820 UP000036403 UP000095300 UP000095301 UP000075809 UP000076502 UP000242457 UP000078492 UP000053825 UP000002320 UP000007819 UP000007266 UP000002358 UP000215335 UP000078200 UP000092445 UP000092444 UP000091820 UP000005203 UP000078541 UP000001070 UP000183832 UP000037069 UP000009192 UP000002282 UP000007801 UP000008744 UP000192221 UP000268350 UP000008792 UP000001819 UP000008711 UP000092553 UP000000304 UP000001292 UP000000803 UP000000305 UP000007798 UP000030742 UP000019118
UP000245037 UP000192223 UP000027135 UP000053105 UP000009046 UP000053097 UP000279307 UP000005205 UP000069940 UP000249989 UP000008237 UP000078540 UP000000311 UP000235965 UP000008820 UP000036403 UP000095300 UP000095301 UP000075809 UP000076502 UP000242457 UP000078492 UP000053825 UP000002320 UP000007819 UP000007266 UP000002358 UP000215335 UP000078200 UP000092445 UP000092444 UP000091820 UP000005203 UP000078541 UP000001070 UP000183832 UP000037069 UP000009192 UP000002282 UP000007801 UP000008744 UP000192221 UP000268350 UP000008792 UP000001819 UP000008711 UP000092553 UP000000304 UP000001292 UP000000803 UP000000305 UP000007798 UP000030742 UP000019118
Interpro
IPR008271
Ser/Thr_kinase_AS
+ More
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
IPR001206 Diacylglycerol_kinase_cat_dom
IPR000756 Diacylglycerol_kin_accessory
IPR037607 DGK
IPR002219 PE/DAG-bd
IPR017438 ATP-NAD_kinase_N
IPR016064 NAD/diacylglycerol_kinase_sf
IPR011990 TPR-like_helical_dom_sf
IPR013026 TPR-contain_dom
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
IPR001206 Diacylglycerol_kinase_cat_dom
IPR000756 Diacylglycerol_kin_accessory
IPR037607 DGK
IPR002219 PE/DAG-bd
IPR017438 ATP-NAD_kinase_N
IPR016064 NAD/diacylglycerol_kinase_sf
IPR011990 TPR-like_helical_dom_sf
IPR013026 TPR-contain_dom
Gene 3D
ProteinModelPortal
H9J7Z9
A0A2A4JEF0
D0AB87
L7WW91
A0A2H1VVK4
A0A212EP69
+ More
A0A194QAR5 A0A194QTH5 S4NSS2 A0A0L7LPN6 A0A1Y1JW72 A0A2P8Y5R8 A0A1W4XB76 A0A1B6IB42 A0A1B6GET5 A0A067QN57 A0A1B6L3U1 A9XK84 A0A0T6AYY0 A0A0M9AAQ5 E0VQ74 A0A026WS53 A0A158NNP5 A0A023ER47 E2B907 A0A151HZL0 A0A1L8DM37 E2AZT9 A0A2J7PXF6 Q17KN4 Q67EX4 U5EW25 A0A069DYN1 A0A1L8DMC4 A0A0J7KV34 A0A0K8TSJ2 A0A0C9QM81 A0A224XTF1 A0A0P4VRJ8 R4G3V7 A0A1I8PC03 A0A1I8M2L4 T1PDH9 A0A151X7T9 A0A1Q3F5W1 A0A154P2K1 A0A2A3E1J1 A0A0V0G9F5 A0A151J3E7 A0A0L7QK85 B0W7R1 V5GF02 J9JYM3 A0A336LUS6 D6WX74 K7J2E0 A0A232F0F6 A0A1A9VNI1 A0A1B0AFB7 D3TLE9 A0A1A9WVR0 A0A088AVZ4 A0A0K8UXF9 A0A034WHR3 A0A2H8TDS5 A0A195FS05 A0A0A1XEX4 A0A310SX15 A0A2R7W8R2 B4JLY8 A0A1J1IV25 A0A0L0BUV4 T1IUF7 B4L621 W8B7L0 B4Q205 B3MS42 B4GW95 A0A1W4VK79 A0A3B0KRW6 B4M728 Q29IB7 B3NX72 A0A0M4F9A4 B4NUD3 B4IGM0 O62602 E9H2U1 Q9U983 B4NEN3 A0A0N8CT73 A0A0N8DCF9 A0A0P5L8K9 A0A0N7ZKQ6 A0A0N8AGI5 A0A0P5Y8M6 U4U2E7 A0A0N8DX38 N6UGG3 A0A0P5EKH4
A0A194QAR5 A0A194QTH5 S4NSS2 A0A0L7LPN6 A0A1Y1JW72 A0A2P8Y5R8 A0A1W4XB76 A0A1B6IB42 A0A1B6GET5 A0A067QN57 A0A1B6L3U1 A9XK84 A0A0T6AYY0 A0A0M9AAQ5 E0VQ74 A0A026WS53 A0A158NNP5 A0A023ER47 E2B907 A0A151HZL0 A0A1L8DM37 E2AZT9 A0A2J7PXF6 Q17KN4 Q67EX4 U5EW25 A0A069DYN1 A0A1L8DMC4 A0A0J7KV34 A0A0K8TSJ2 A0A0C9QM81 A0A224XTF1 A0A0P4VRJ8 R4G3V7 A0A1I8PC03 A0A1I8M2L4 T1PDH9 A0A151X7T9 A0A1Q3F5W1 A0A154P2K1 A0A2A3E1J1 A0A0V0G9F5 A0A151J3E7 A0A0L7QK85 B0W7R1 V5GF02 J9JYM3 A0A336LUS6 D6WX74 K7J2E0 A0A232F0F6 A0A1A9VNI1 A0A1B0AFB7 D3TLE9 A0A1A9WVR0 A0A088AVZ4 A0A0K8UXF9 A0A034WHR3 A0A2H8TDS5 A0A195FS05 A0A0A1XEX4 A0A310SX15 A0A2R7W8R2 B4JLY8 A0A1J1IV25 A0A0L0BUV4 T1IUF7 B4L621 W8B7L0 B4Q205 B3MS42 B4GW95 A0A1W4VK79 A0A3B0KRW6 B4M728 Q29IB7 B3NX72 A0A0M4F9A4 B4NUD3 B4IGM0 O62602 E9H2U1 Q9U983 B4NEN3 A0A0N8CT73 A0A0N8DCF9 A0A0P5L8K9 A0A0N7ZKQ6 A0A0N8AGI5 A0A0P5Y8M6 U4U2E7 A0A0N8DX38 N6UGG3 A0A0P5EKH4
PDB
3VN9
E-value=5.15126e-98,
Score=912
Ontologies
GO
GO:0005524
GO:0004674
GO:0004672
GO:0000187
GO:0005737
GO:0004708
GO:0023014
GO:0032147
GO:0031098
GO:0035556
GO:0003951
GO:0007205
GO:0004143
GO:0038066
GO:0031435
GO:0040018
GO:0090090
GO:0002385
GO:0048082
GO:0007309
GO:0045793
GO:0001934
GO:0004697
GO:0000165
GO:0007314
GO:0006468
GO:0016021
GO:0004535
GO:0003723
GO:0005634
GO:0006402
GO:0046872
GO:0003824
PANTHER
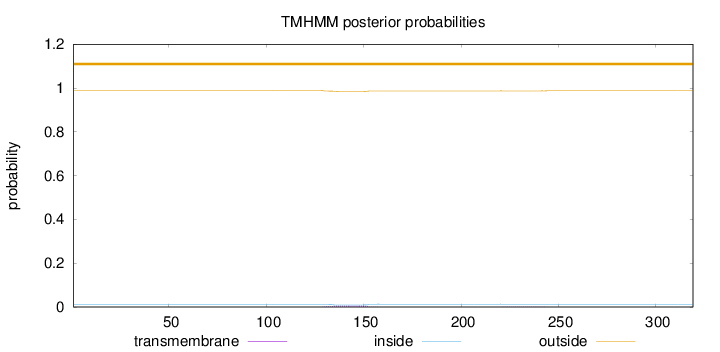
Topology
Length:
319
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14094
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01173
outside
1 - 319
Population Genetic Test Statistics
Pi
314.532692
Theta
166.358616
Tajima's D
2.451871
CLR
116.550715
CSRT
0.941402929853507
Interpretation
Uncertain
