Gene
KWMTBOMO10136
Pre Gene Modal
BGIBMGA005562
Annotation
PREDICTED:_uncharacterized_protein_KIAA0195?_partial_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.79
Sequence
CDS
ATGTGCGAGACGCCTTACTTGAAGAATCTAAAGATGGCACTCGAACAAGCAACAACCAGGCCGGTCACTGTTTTTGAGAAACAAAGGTATTTGTGCACAGTGAAGGTGATGGAAGGCATAGTGCTGCCCCTGGTCATATCCTTAGTTGTGGTGGCGAGCTTCCTGCGTCACGTGTACCATTTGCCCGGCGTGACCCACTGGACGGAGATATACTTCCTCCAGACCATAGCCGCTTCACTGCCCCTATTGCAAATCGTGTTCCCCATAGCTTGGGTCACGCTTAACTGTTACGGATTGGCGAGATTTAAGCAAATAGCAGAAACCCGCCAAAAATACGTCCGTCCGAAGTCATGTTCCATATCAGAGGACGCGAGCGACCCTCTGCTTCAAGCACTGAGCGAATCCAGTCTGAAACCGGAGAGCATCAAGTCATTAGCAAAGACATTCTTCAACATCTTGACTGGTAGAGAAGACATGGTATCTAGGACTGCCAATATCGTACATGTCCTGGGATCTTTGTCTGCTCTATCGTGTGTGGACAAAAAGGGGATATTATCATGGCCAAATCCGACACCAGAGAAAGTGTTTTTCCTTCGTAATTCGGATCCCACGTCGCACAACTCGAGCAAAACTAGTCTGGCCGGCTCCCTGGGTCATCCCGGAGATTCGGTACTGGAACAGCCAGATAACAGGCACCACGAACCGTCTACTGTTGTCGAGGTGTTAGACCTGACCCACGACCAGAACGCGCCGTTCTGCGTGGAGTTCGACGACCAGCGCTGGCGGCGGCACCTGGCGCTGCTGAAGCCGCTGGGGCTGGCGGCGCTGCTCAACACGTGCGCGCCCGCCCCCAGACACACGTATGCGCACTTCTGCGCGCACCTCACCTGCGAAGCTCAGTTGAGGTACGATTATTTTTATTGCTTAGATGTGTGGACGACCTCACAGCCCACCTGGTGTTAA
Protein
MCETPYLKNLKMALEQATTRPVTVFEKQRYLCTVKVMEGIVLPLVISLVVVASFLRHVYHLPGVTHWTEIYFLQTIAASLPLLQIVFPIAWVTLNCYGLARFKQIAETRQKYVRPKSCSISEDASDPLLQALSESSLKPESIKSLAKTFFNILTGREDMVSRTANIVHVLGSLSALSCVDKKGILSWPNPTPEKVFFLRNSDPTSHNSSKTSLAGSLGHPGDSVLEQPDNRHHEPSTVVEVLDLTHDQNAPFCVEFDDQRWRRHLALLKPLGLAALLNTCAPAPRHTYAHFCAHLTCEAQLRYDYFYCLDVWTTSQPTWC
Summary
Uniprot
A0A2H1W431
A0A2H1W436
H9J7S0
A0A2A4JEV8
A0A194QB23
A0A194QSL0
+ More
A0A3S2L8H5 L7X3W0 D0AB86 A0A212EPB0 A0A1E1WS86 A0A0C9Q622 A0A0C9RD07 A0A0C9QMF3 A0A0C9PL89 A0A0C9RMK5 A0A151J7P1 A0A151JXP4 E2B474 A0A310SMW2 A0A232ER02 A0A2A3EAG4 F4WHR9 A0A195B5D9 A0A158NM24 E1ZV00 A0A026WG49 A0A151XIG3 A0A0L7QQV6 A0A3L8DJS8 A0A087ZXX4 A0A0N0U4H9 A0A195CWZ6 A0A1Y1JYP7 A0A1Y1JTF6 A0A1Y1JWG9 A0A1Y1JYK5 A0A1Y1JTH2 A0A1Y1JWI3 A0A1B6CFK5 A0A1B6C8E3 A0A1Y1JTV9 A0A1Y1JWC3 A0A1B6DZA4 A0A1B6CRC8 A0A1B6D8B3 A0A1B6DHS1 A0A1B6BYR1 A0A1B6DTF3 A0A1B6CCC8 A0A1B6BXT8 A0A1B6EF44 A0A1B6DN27 D6WX73 A0A3B0K8M9 A0A0R3NT85 Q29LJ3 A0A0R3NYJ3 A0A1B6GK17 A0A1W4WA34 A0A1W4W971 A0A1B6GU86 A0A0R1DJ30 B4NYJ8 A0A0R1DJ53 A0A067QXM3 B4MZM4 A0A0J9QVI0 A0A0J9QVJ3 B4I385 B3N3R6 A0A0Q5WID4 A0A0P8XY64 A0A0P8ZI66 A0A0J9QVM0 B4QA67 B3MU39 A0A0Q5WLE1 A0A0P8XHV7 A0A0P9BRS4 Q7KU45 Q9VQZ1 A0A0N8ES84 A0A1B6HB37 A0A0M4EBF0 A0A182GSD3 A0A1B6JB83 A0A1B6HLI4 N6UJ90 U4UKU9 A0A1B6JBX6 E0VQ73 B4KES2 A0A0Q9XEQ8 A0A0Q9XIR8 W8B4C2 A0A1A9WGE0 A0A182H3K9 W8AHZ9
A0A3S2L8H5 L7X3W0 D0AB86 A0A212EPB0 A0A1E1WS86 A0A0C9Q622 A0A0C9RD07 A0A0C9QMF3 A0A0C9PL89 A0A0C9RMK5 A0A151J7P1 A0A151JXP4 E2B474 A0A310SMW2 A0A232ER02 A0A2A3EAG4 F4WHR9 A0A195B5D9 A0A158NM24 E1ZV00 A0A026WG49 A0A151XIG3 A0A0L7QQV6 A0A3L8DJS8 A0A087ZXX4 A0A0N0U4H9 A0A195CWZ6 A0A1Y1JYP7 A0A1Y1JTF6 A0A1Y1JWG9 A0A1Y1JYK5 A0A1Y1JTH2 A0A1Y1JWI3 A0A1B6CFK5 A0A1B6C8E3 A0A1Y1JTV9 A0A1Y1JWC3 A0A1B6DZA4 A0A1B6CRC8 A0A1B6D8B3 A0A1B6DHS1 A0A1B6BYR1 A0A1B6DTF3 A0A1B6CCC8 A0A1B6BXT8 A0A1B6EF44 A0A1B6DN27 D6WX73 A0A3B0K8M9 A0A0R3NT85 Q29LJ3 A0A0R3NYJ3 A0A1B6GK17 A0A1W4WA34 A0A1W4W971 A0A1B6GU86 A0A0R1DJ30 B4NYJ8 A0A0R1DJ53 A0A067QXM3 B4MZM4 A0A0J9QVI0 A0A0J9QVJ3 B4I385 B3N3R6 A0A0Q5WID4 A0A0P8XY64 A0A0P8ZI66 A0A0J9QVM0 B4QA67 B3MU39 A0A0Q5WLE1 A0A0P8XHV7 A0A0P9BRS4 Q7KU45 Q9VQZ1 A0A0N8ES84 A0A1B6HB37 A0A0M4EBF0 A0A182GSD3 A0A1B6JB83 A0A1B6HLI4 N6UJ90 U4UKU9 A0A1B6JBX6 E0VQ73 B4KES2 A0A0Q9XEQ8 A0A0Q9XIR8 W8B4C2 A0A1A9WGE0 A0A182H3K9 W8AHZ9
Pubmed
19121390
26354079
23674305
22118469
20798317
28648823
+ More
21719571 21347285 24508170 30249741 28004739 18362917 19820115 15632085 17994087 17550304 24845553 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26999592 26483478 23537049 20566863 24495485
21719571 21347285 24508170 30249741 28004739 18362917 19820115 15632085 17994087 17550304 24845553 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26999592 26483478 23537049 20566863 24495485
EMBL
ODYU01006205
SOQ47850.1
ODYU01006206
SOQ47851.1
BABH01020203
BABH01020204
+ More
BABH01020205 BABH01020206 NWSH01001832 PCG69963.1 KQ459232 KPJ02629.1 KQ461155 KPJ08304.1 RSAL01000090 RVE48102.1 KC469893 AGC92690.2 FP102340 CBH09281.1 AGBW02013517 OWR43291.1 GDQN01001358 JAT89696.1 GBYB01009583 JAG79350.1 GBYB01011032 JAG80799.1 GBYB01001767 JAG71534.1 GBYB01001768 JAG71535.1 GBYB01009580 JAG79347.1 KQ979658 KYN19892.1 KQ981561 KYN39827.1 GL445515 EFN89491.1 KQ759847 OAD62585.1 NNAY01002685 OXU20778.1 KZ288308 PBC28715.1 GL888167 EGI66276.1 KQ976604 KYM79404.1 ADTU01020204 GL434335 EFN74998.1 KK107238 EZA54621.1 KQ982109 KYQ60000.1 KQ414784 KOC61013.1 QOIP01000007 RLU20670.1 KQ435834 KOX71551.1 KQ977171 KYN05181.1 GEZM01101564 GEZM01101559 GEZM01101557 JAV52545.1 GEZM01101544 JAV52589.1 GEZM01101577 GEZM01101572 GEZM01101571 GEZM01101563 GEZM01101562 GEZM01101561 GEZM01101555 GEZM01101554 GEZM01101552 GEZM01101543 GEZM01101542 JAV52531.1 GEZM01101574 JAV52505.1 GEZM01101579 GEZM01101578 GEZM01101575 GEZM01101568 GEZM01101560 GEZM01101558 GEZM01101551 GEZM01101550 GEZM01101548 GEZM01101546 GEZM01101541 GEZM01101540 JAV52604.1 GEZM01101545 JAV52588.1 GEDC01025148 JAS12150.1 GEDC01027521 JAS09777.1 GEZM01101567 GEZM01101556 JAV52523.1 GEZM01101576 GEZM01101573 GEZM01101570 GEZM01101569 GEZM01101566 GEZM01101565 GEZM01101553 GEZM01101549 GEZM01101547 JAV52508.1 GEDC01006317 JAS30981.1 GEDC01024062 GEDC01021433 JAS13236.1 JAS15865.1 GEDC01015412 JAS21886.1 GEDC01020970 GEDC01012123 JAS16328.1 JAS25175.1 GEDC01030912 GEDC01010908 JAS06386.1 JAS26390.1 GEDC01029363 GEDC01008363 JAS07935.1 JAS28935.1 GEDC01026348 JAS10950.1 GEDC01031434 GEDC01026722 JAS05864.1 JAS10576.1 GEDC01000814 JAS36484.1 GEDC01023274 GEDC01010258 JAS14024.1 JAS27040.1 KQ971361 EFA08800.1 OUUW01000004 SPP79888.1 CH379060 KRT04430.1 EAL34052.2 KRT04431.1 GECZ01007039 JAS62730.1 GECZ01003767 JAS66002.1 CM000157 KRJ97269.1 EDW87631.1 KRJ97268.1 KK853134 KDR10862.1 CH963920 EDW77809.1 CM002910 KMY88057.1 KMY88058.1 CH480820 EDW54230.1 CH954177 EDV57725.1 KQS70089.1 CH902624 KPU74432.1 KPU74431.1 KMY88056.1 CM000361 EDX03705.1 EDV33368.1 KQS70090.1 KPU74433.1 KPU74430.1 AE014134 AAN10345.2 AGB92559.1 BT015990 AAF51018.3 AAV36875.1 GDUN01000178 JAN95741.1 GECU01035785 JAS71921.1 CP012523 ALC38747.1 JXUM01084398 KQ563435 KXJ73833.1 GECU01011423 JAS96283.1 GECU01032144 JAS75562.1 APGK01022082 KB740398 ENN80731.1 KB632255 ERL90630.1 GECU01011041 JAS96665.1 DS235392 EEB15529.1 CH933807 EDW12972.1 KRG03524.1 KRG03523.1 GAMC01018439 JAB88116.1 JXUM01107644 JXUM01107645 JXUM01107646 KQ565160 KXJ71272.1 GAMC01018440 JAB88115.1
BABH01020205 BABH01020206 NWSH01001832 PCG69963.1 KQ459232 KPJ02629.1 KQ461155 KPJ08304.1 RSAL01000090 RVE48102.1 KC469893 AGC92690.2 FP102340 CBH09281.1 AGBW02013517 OWR43291.1 GDQN01001358 JAT89696.1 GBYB01009583 JAG79350.1 GBYB01011032 JAG80799.1 GBYB01001767 JAG71534.1 GBYB01001768 JAG71535.1 GBYB01009580 JAG79347.1 KQ979658 KYN19892.1 KQ981561 KYN39827.1 GL445515 EFN89491.1 KQ759847 OAD62585.1 NNAY01002685 OXU20778.1 KZ288308 PBC28715.1 GL888167 EGI66276.1 KQ976604 KYM79404.1 ADTU01020204 GL434335 EFN74998.1 KK107238 EZA54621.1 KQ982109 KYQ60000.1 KQ414784 KOC61013.1 QOIP01000007 RLU20670.1 KQ435834 KOX71551.1 KQ977171 KYN05181.1 GEZM01101564 GEZM01101559 GEZM01101557 JAV52545.1 GEZM01101544 JAV52589.1 GEZM01101577 GEZM01101572 GEZM01101571 GEZM01101563 GEZM01101562 GEZM01101561 GEZM01101555 GEZM01101554 GEZM01101552 GEZM01101543 GEZM01101542 JAV52531.1 GEZM01101574 JAV52505.1 GEZM01101579 GEZM01101578 GEZM01101575 GEZM01101568 GEZM01101560 GEZM01101558 GEZM01101551 GEZM01101550 GEZM01101548 GEZM01101546 GEZM01101541 GEZM01101540 JAV52604.1 GEZM01101545 JAV52588.1 GEDC01025148 JAS12150.1 GEDC01027521 JAS09777.1 GEZM01101567 GEZM01101556 JAV52523.1 GEZM01101576 GEZM01101573 GEZM01101570 GEZM01101569 GEZM01101566 GEZM01101565 GEZM01101553 GEZM01101549 GEZM01101547 JAV52508.1 GEDC01006317 JAS30981.1 GEDC01024062 GEDC01021433 JAS13236.1 JAS15865.1 GEDC01015412 JAS21886.1 GEDC01020970 GEDC01012123 JAS16328.1 JAS25175.1 GEDC01030912 GEDC01010908 JAS06386.1 JAS26390.1 GEDC01029363 GEDC01008363 JAS07935.1 JAS28935.1 GEDC01026348 JAS10950.1 GEDC01031434 GEDC01026722 JAS05864.1 JAS10576.1 GEDC01000814 JAS36484.1 GEDC01023274 GEDC01010258 JAS14024.1 JAS27040.1 KQ971361 EFA08800.1 OUUW01000004 SPP79888.1 CH379060 KRT04430.1 EAL34052.2 KRT04431.1 GECZ01007039 JAS62730.1 GECZ01003767 JAS66002.1 CM000157 KRJ97269.1 EDW87631.1 KRJ97268.1 KK853134 KDR10862.1 CH963920 EDW77809.1 CM002910 KMY88057.1 KMY88058.1 CH480820 EDW54230.1 CH954177 EDV57725.1 KQS70089.1 CH902624 KPU74432.1 KPU74431.1 KMY88056.1 CM000361 EDX03705.1 EDV33368.1 KQS70090.1 KPU74433.1 KPU74430.1 AE014134 AAN10345.2 AGB92559.1 BT015990 AAF51018.3 AAV36875.1 GDUN01000178 JAN95741.1 GECU01035785 JAS71921.1 CP012523 ALC38747.1 JXUM01084398 KQ563435 KXJ73833.1 GECU01011423 JAS96283.1 GECU01032144 JAS75562.1 APGK01022082 KB740398 ENN80731.1 KB632255 ERL90630.1 GECU01011041 JAS96665.1 DS235392 EEB15529.1 CH933807 EDW12972.1 KRG03524.1 KRG03523.1 GAMC01018439 JAB88116.1 JXUM01107644 JXUM01107645 JXUM01107646 KQ565160 KXJ71272.1 GAMC01018440 JAB88115.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000078492 UP000078541 UP000008237 UP000215335 UP000242457 UP000007755 UP000078540 UP000005205 UP000000311 UP000053097 UP000075809 UP000053825 UP000279307 UP000005203 UP000053105 UP000078542 UP000007266 UP000268350 UP000001819 UP000192221 UP000002282 UP000027135 UP000007798 UP000001292 UP000008711 UP000007801 UP000000304 UP000000803 UP000092553 UP000069940 UP000249989 UP000019118 UP000030742 UP000009046 UP000009192 UP000091820
UP000078492 UP000078541 UP000008237 UP000215335 UP000242457 UP000007755 UP000078540 UP000005205 UP000000311 UP000053097 UP000075809 UP000053825 UP000279307 UP000005203 UP000053105 UP000078542 UP000007266 UP000268350 UP000001819 UP000192221 UP000002282 UP000027135 UP000007798 UP000001292 UP000008711 UP000007801 UP000000304 UP000000803 UP000092553 UP000069940 UP000249989 UP000019118 UP000030742 UP000009046 UP000009192 UP000091820
Interpro
Gene 3D
ProteinModelPortal
A0A2H1W431
A0A2H1W436
H9J7S0
A0A2A4JEV8
A0A194QB23
A0A194QSL0
+ More
A0A3S2L8H5 L7X3W0 D0AB86 A0A212EPB0 A0A1E1WS86 A0A0C9Q622 A0A0C9RD07 A0A0C9QMF3 A0A0C9PL89 A0A0C9RMK5 A0A151J7P1 A0A151JXP4 E2B474 A0A310SMW2 A0A232ER02 A0A2A3EAG4 F4WHR9 A0A195B5D9 A0A158NM24 E1ZV00 A0A026WG49 A0A151XIG3 A0A0L7QQV6 A0A3L8DJS8 A0A087ZXX4 A0A0N0U4H9 A0A195CWZ6 A0A1Y1JYP7 A0A1Y1JTF6 A0A1Y1JWG9 A0A1Y1JYK5 A0A1Y1JTH2 A0A1Y1JWI3 A0A1B6CFK5 A0A1B6C8E3 A0A1Y1JTV9 A0A1Y1JWC3 A0A1B6DZA4 A0A1B6CRC8 A0A1B6D8B3 A0A1B6DHS1 A0A1B6BYR1 A0A1B6DTF3 A0A1B6CCC8 A0A1B6BXT8 A0A1B6EF44 A0A1B6DN27 D6WX73 A0A3B0K8M9 A0A0R3NT85 Q29LJ3 A0A0R3NYJ3 A0A1B6GK17 A0A1W4WA34 A0A1W4W971 A0A1B6GU86 A0A0R1DJ30 B4NYJ8 A0A0R1DJ53 A0A067QXM3 B4MZM4 A0A0J9QVI0 A0A0J9QVJ3 B4I385 B3N3R6 A0A0Q5WID4 A0A0P8XY64 A0A0P8ZI66 A0A0J9QVM0 B4QA67 B3MU39 A0A0Q5WLE1 A0A0P8XHV7 A0A0P9BRS4 Q7KU45 Q9VQZ1 A0A0N8ES84 A0A1B6HB37 A0A0M4EBF0 A0A182GSD3 A0A1B6JB83 A0A1B6HLI4 N6UJ90 U4UKU9 A0A1B6JBX6 E0VQ73 B4KES2 A0A0Q9XEQ8 A0A0Q9XIR8 W8B4C2 A0A1A9WGE0 A0A182H3K9 W8AHZ9
A0A3S2L8H5 L7X3W0 D0AB86 A0A212EPB0 A0A1E1WS86 A0A0C9Q622 A0A0C9RD07 A0A0C9QMF3 A0A0C9PL89 A0A0C9RMK5 A0A151J7P1 A0A151JXP4 E2B474 A0A310SMW2 A0A232ER02 A0A2A3EAG4 F4WHR9 A0A195B5D9 A0A158NM24 E1ZV00 A0A026WG49 A0A151XIG3 A0A0L7QQV6 A0A3L8DJS8 A0A087ZXX4 A0A0N0U4H9 A0A195CWZ6 A0A1Y1JYP7 A0A1Y1JTF6 A0A1Y1JWG9 A0A1Y1JYK5 A0A1Y1JTH2 A0A1Y1JWI3 A0A1B6CFK5 A0A1B6C8E3 A0A1Y1JTV9 A0A1Y1JWC3 A0A1B6DZA4 A0A1B6CRC8 A0A1B6D8B3 A0A1B6DHS1 A0A1B6BYR1 A0A1B6DTF3 A0A1B6CCC8 A0A1B6BXT8 A0A1B6EF44 A0A1B6DN27 D6WX73 A0A3B0K8M9 A0A0R3NT85 Q29LJ3 A0A0R3NYJ3 A0A1B6GK17 A0A1W4WA34 A0A1W4W971 A0A1B6GU86 A0A0R1DJ30 B4NYJ8 A0A0R1DJ53 A0A067QXM3 B4MZM4 A0A0J9QVI0 A0A0J9QVJ3 B4I385 B3N3R6 A0A0Q5WID4 A0A0P8XY64 A0A0P8ZI66 A0A0J9QVM0 B4QA67 B3MU39 A0A0Q5WLE1 A0A0P8XHV7 A0A0P9BRS4 Q7KU45 Q9VQZ1 A0A0N8ES84 A0A1B6HB37 A0A0M4EBF0 A0A182GSD3 A0A1B6JB83 A0A1B6HLI4 N6UJ90 U4UKU9 A0A1B6JBX6 E0VQ73 B4KES2 A0A0Q9XEQ8 A0A0Q9XIR8 W8B4C2 A0A1A9WGE0 A0A182H3K9 W8AHZ9
Ontologies
PANTHER
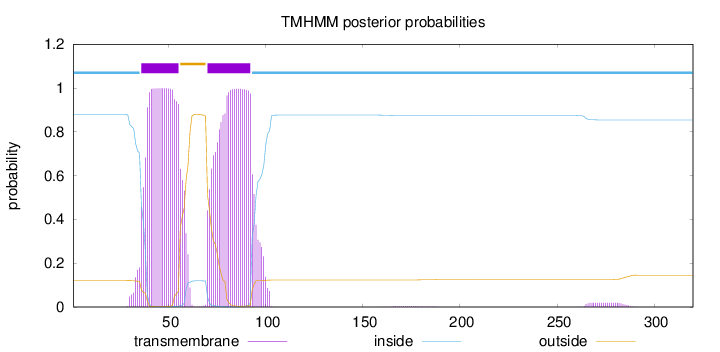
Topology
Length:
320
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.8484300000001
Exp number, first 60 AAs:
21.32979
Total prob of N-in:
0.87910
POSSIBLE N-term signal
sequence
inside
1 - 35
TMhelix
36 - 55
outside
56 - 69
TMhelix
70 - 92
inside
93 - 320
Population Genetic Test Statistics
Pi
209.433747
Theta
146.407414
Tajima's D
2.054375
CLR
19.267345
CSRT
0.892405379731013
Interpretation
Uncertain
