Gene
KWMTBOMO10129 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005560
Annotation
proteasome_26S_non-ATPase_subunit_4_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.259
Sequence
CDS
ATGGTCTTAGAAAGTACTATGATCTGTGTGGACAATAGTGACTACATGAGAAATGGAGATTTCTTGCCGACACGACTGCAAGCTCAGCAGGATGCTGTTAATTTGGTATGTCATTCCAAAACACGATCCAATCCTGAGAATAATGTCGGATTATTAACTCTTGCAAACGTCGAAGTATTAGCCACACTTACTAGCGACGTAGGACGTATCATGTCCAAGCTCCATCGCGTCCAGCCAAACGGTGACATCAATCTATTGACCGGAATCCGCATTGCACATTTAGCTTTAAAACACCGTCAGGGAAAGAATCACAAGATGCGCATAGTAGTTTTTGTCGGTTCCCCTGTCAACACTGACGAAAAGGAACTCGTAAAACTAGCTAAAAGGCTAAAGAAGGAGAAAGTCAACTGTGATGTTGTTTCGTTTGGAGAGGATTCAGAAAACAACCCCCTTTTGACTACCTTTGTCAACACACTAAATGGGAAAGACACATCGACTGGGGGAAGTCACCTTGTGTCTGTGCCAGCTGGAGGCTGTGTTGTGCTTTCTGAGGCTTTGATAACAAGTCCTCTGATAGGTGGTGATGGAGCAGGCCCTTCTGGCTCTGGTTTGTCTCCTTTCGAATTTGGAGTAGATCCCAATGTAGACCCAGAGCTCGCATTAGCTCTACGTGTGTCGATGGAAGAACAAAGGCAACGTCAAGAAGAAGAGTCTCGTCGTCAACAGACCACAACTGAAGGCGAAACTGGCACTGCCACTGAAGGATCAGATACAGCAGTAGAGAGAGCCATTGCTATGTCTCTCGGTCGTGATGCAATGGAACTCTCTGAAGAGGAGCAGATTGCGCTTGCAATGCAAATGAGCATGCAACAAGAAGCACCCCAGGCTGAAGAGAGCATGGATGTTTCTGAAGAATATGCAGAAGTTATGAATGATCCAGCATTTTTACAGAGTGTGCTTGAGAACCTTCCCGGAGTAGACCCACAGAGTGAAGCCATCAGAAATGCTATGTCAACAATCAAGAAGGATAAGGATGAGAAGGATGAGAAAGACAAAGACGGAAAAGGATCCACATCTAAAGAGAATTAG
Protein
MVLESTMICVDNSDYMRNGDFLPTRLQAQQDAVNLVCHSKTRSNPENNVGLLTLANVEVLATLTSDVGRIMSKLHRVQPNGDINLLTGIRIAHLALKHRQGKNHKMRIVVFVGSPVNTDEKELVKLAKRLKKEKVNCDVVSFGEDSENNPLLTTFVNTLNGKDTSTGGSHLVSVPAGGCVVLSEALITSPLIGGDGAGPSGSGLSPFEFGVDPNVDPELALALRVSMEEQRQRQEEESRRQQTTTEGETGTATEGSDTAVERAIAMSLGRDAMELSEEEQIALAMQMSMQQEAPQAEESMDVSEEYAEVMNDPAFLQSVLENLPGVDPQSEAIRNAMSTIKKDKDEKDEKDKDGKGSTSKEN
Summary
Uniprot
Q2F5X3
D0AB76
L7X3X7
A0A212FKP4
A0A2A4JIA6
A0A2H1X328
+ More
S4P8H2 A0A194QXK8 A0A194QAX9 E3UKM5 A0A0L7LIK5 D8VIK0 D8VIM0 D8VII0 D8VIK9 D8VIJ9 D8VII1 D8VIK1 G9JQN6 G9JPI5 G9JQQ4 G9JQN1 G9JQT3 G9JQN8 G9JQQ9 G9JQM2 G9JPG9 G9JPE6 G9JPH2 G9JPE0 G9JPG6 G9JPF4 G9JPD8 G9JPE7 G9JPF5 G9JPC3 G9JPC8 G9JPG1 G9JPH5 G9JPI7 G9JPB3 G9JPH0 G9JPH4 G9JPB8 G9JPH3 G9JQP1 G9JQN4 G9JQQ7 G9JQN9 G9JQN3 G9JQP2 G9JQM7 G9JQQ1 G9JQN5 G9JQR4 G9JQS3 G9JQS7 G9JQP7 G9JQT2 G9JQM5 G9JPB0 G9JPB6 G9JQQ6 G9JQP0 G9JQR8 G9JPG7 G9JQP3 G9JPB5 G9JPD4 G9JPF3 G9JPD0 G9JQQ0 G9JPF6 G9JPD9 G9JPD5 G9JQR1 G9JQQ5 G9JPE1 G9JQM3 R4WDF5 A0A0P4VUT1 A0A0V0G6L2 R4FQB0 A0A224XRH9 A0A1B6CQ59 A0A1S3H8E6 A0A1S3H700 S7Q067 G1PRQ7 A0A151M585 L5K2W8 T1D9J7
S4P8H2 A0A194QXK8 A0A194QAX9 E3UKM5 A0A0L7LIK5 D8VIK0 D8VIM0 D8VII0 D8VIK9 D8VIJ9 D8VII1 D8VIK1 G9JQN6 G9JPI5 G9JQQ4 G9JQN1 G9JQT3 G9JQN8 G9JQQ9 G9JQM2 G9JPG9 G9JPE6 G9JPH2 G9JPE0 G9JPG6 G9JPF4 G9JPD8 G9JPE7 G9JPF5 G9JPC3 G9JPC8 G9JPG1 G9JPH5 G9JPI7 G9JPB3 G9JPH0 G9JPH4 G9JPB8 G9JPH3 G9JQP1 G9JQN4 G9JQQ7 G9JQN9 G9JQN3 G9JQP2 G9JQM7 G9JQQ1 G9JQN5 G9JQR4 G9JQS3 G9JQS7 G9JQP7 G9JQT2 G9JQM5 G9JPB0 G9JPB6 G9JQQ6 G9JQP0 G9JQR8 G9JPG7 G9JQP3 G9JPB5 G9JPD4 G9JPF3 G9JPD0 G9JQQ0 G9JPF6 G9JPD9 G9JPD5 G9JQR1 G9JQQ5 G9JPE1 G9JQM3 R4WDF5 A0A0P4VUT1 A0A0V0G6L2 R4FQB0 A0A224XRH9 A0A1B6CQ59 A0A1S3H8E6 A0A1S3H700 S7Q067 G1PRQ7 A0A151M585 L5K2W8 T1D9J7
Pubmed
EMBL
BABH01020216
DQ311300
ABD36244.1
FP102339
CBH09271.1
KC469893
+ More
AGC92710.1 AGBW02008005 OWR54313.1 NWSH01001365 PCG71529.1 ODYU01013053 SOQ59709.1 GAIX01005931 JAA86629.1 KQ461155 KPJ08311.1 KQ459232 KPJ02622.1 HM449899 ADO33034.1 JTDY01000993 KOB75174.1 GQ986847 GQ986849 GQ986850 GQ986851 GQ986852 GQ986853 GQ986854 GQ986855 GQ986857 GQ986859 GQ986860 GQ986861 GQ986862 GQ986863 GQ986864 GQ986865 GQ986866 ADI80996.1 GQ986867 GQ986868 GQ986869 GQ986870 GQ986871 GQ986872 GQ986873 GQ986874 GQ986875 GQ986876 GQ986877 GQ986878 GQ986879 GQ986880 GQ986881 GQ986882 GQ986883 GQ986884 GQ986885 GQ986886 ADI81016.1 GQ986827 GQ986839 GQ986840 GQ986843 GQ986845 ADI80976.1 GQ986856 GQ986858 ADI81005.1 GQ986846 ADI80995.1 GQ986828 GQ986829 GQ986830 GQ986831 GQ986832 GQ986833 GQ986834 GQ986835 GQ986836 GQ986837 GQ986838 GQ986841 GQ986842 GQ986844 ADI80977.1 GQ986848 ADI80997.1 JN898017 AET96670.1 JN897616 AET96269.1 JN898035 AET96688.1 JN898012 AET96665.1 JN898064 AET96717.1 JN898019 AET96672.1 JN898010 JN898039 JN898040 JN898041 AET96693.1 JN898002 JN898003 JN898005 AET96656.1 JN897600 AET96253.1 JN897577 AET96230.1 JN897603 AET96256.1 JN897571 AET96224.1 JN897594 JN897595 JN897596 JN897597 AET96250.1 JN897582 JN897583 JN897585 AET96238.1 JN897569 JN897573 JN897574 JN897575 AET96222.1 JN897576 JN897578 JN897579 JN897580 AET96231.1 JN897586 JN897588 AET96239.1 JN897553 JN897554 JN897567 JN897568 AET96207.1 JN897552 JN897555 JN897556 JN897557 JN897558 JN897559 JN897560 JN897562 JN897563 JN897564 JN897581 JN897611 JN897612 JN897613 JN897614 AET96212.1 JN897589 JN897590 JN897591 JN897592 JN897593 AET96245.1 JN897606 JN897607 JN897610 AET96259.1 JN897545 JN897615 JN897617 JN897618 AET96271.1 JN897542 JN897544 AET96197.1 JN897599 JN897601 JN897602 AET96254.1 JN897605 JN897608 JN897609 AET96258.1 JN897548 JN897549 JN897550 JN897551 AET96202.1 JN897604 AET96257.1 JN898022 AET96675.1 JN898015 AET96668.1 JN898038 AET96691.1 JN898020 AET96673.1 JN898009 JN898014 AET96667.1 JN898023 AET96676.1 JN898008 AET96661.1 JN898030 JN898032 JN898033 JN898034 AET96685.1 JN898016 JN898018 AET96669.1 JN898045 JN898046 JN898047 JN898048 AET96698.1 JN898043 JN898044 JN898050 JN898051 JN898052 JN898053 JN898054 AET96707.1 JN898055 JN898056 JN898057 JN898058 JN898059 AET96711.1 JN898025 JN898026 JN898027 JN898028 JN898029 AET96681.1 JN898011 JN898013 JN898060 JN898061 JN898062 JN898063 AET96716.1 JN898006 JN898007 AET96659.1 JN897541 AET96194.1 JN897547 AET96200.1 JN898037 AET96690.1 JN898021 AET96674.1 JN898049 AET96702.1 JN897598 AET96251.1 JN898024 AET96677.1 JN897546 AET96199.1 JN897565 AET96218.1 JN897584 AET96237.1 JN897561 AET96214.1 JN898031 AET96684.1 JN897587 AET96240.1 JN897570 AET96223.1 JN897566 AET96219.1 JN898042 AET96695.1 JN898036 AET96689.1 JN897572 AET96225.1 JN898004 AET96657.1 AK417638 BAN20853.1 GDKW01000104 JAI56491.1 GECL01003165 JAP02959.1 GAHY01000504 JAA77006.1 GFTR01005673 JAW10753.1 GEDC01021813 JAS15485.1 KE164279 EPQ16653.1 AAPE02030108 AKHW03006582 KYO19679.1 KB031037 ELK06049.1 GAAZ01001934 JAA96009.1
AGC92710.1 AGBW02008005 OWR54313.1 NWSH01001365 PCG71529.1 ODYU01013053 SOQ59709.1 GAIX01005931 JAA86629.1 KQ461155 KPJ08311.1 KQ459232 KPJ02622.1 HM449899 ADO33034.1 JTDY01000993 KOB75174.1 GQ986847 GQ986849 GQ986850 GQ986851 GQ986852 GQ986853 GQ986854 GQ986855 GQ986857 GQ986859 GQ986860 GQ986861 GQ986862 GQ986863 GQ986864 GQ986865 GQ986866 ADI80996.1 GQ986867 GQ986868 GQ986869 GQ986870 GQ986871 GQ986872 GQ986873 GQ986874 GQ986875 GQ986876 GQ986877 GQ986878 GQ986879 GQ986880 GQ986881 GQ986882 GQ986883 GQ986884 GQ986885 GQ986886 ADI81016.1 GQ986827 GQ986839 GQ986840 GQ986843 GQ986845 ADI80976.1 GQ986856 GQ986858 ADI81005.1 GQ986846 ADI80995.1 GQ986828 GQ986829 GQ986830 GQ986831 GQ986832 GQ986833 GQ986834 GQ986835 GQ986836 GQ986837 GQ986838 GQ986841 GQ986842 GQ986844 ADI80977.1 GQ986848 ADI80997.1 JN898017 AET96670.1 JN897616 AET96269.1 JN898035 AET96688.1 JN898012 AET96665.1 JN898064 AET96717.1 JN898019 AET96672.1 JN898010 JN898039 JN898040 JN898041 AET96693.1 JN898002 JN898003 JN898005 AET96656.1 JN897600 AET96253.1 JN897577 AET96230.1 JN897603 AET96256.1 JN897571 AET96224.1 JN897594 JN897595 JN897596 JN897597 AET96250.1 JN897582 JN897583 JN897585 AET96238.1 JN897569 JN897573 JN897574 JN897575 AET96222.1 JN897576 JN897578 JN897579 JN897580 AET96231.1 JN897586 JN897588 AET96239.1 JN897553 JN897554 JN897567 JN897568 AET96207.1 JN897552 JN897555 JN897556 JN897557 JN897558 JN897559 JN897560 JN897562 JN897563 JN897564 JN897581 JN897611 JN897612 JN897613 JN897614 AET96212.1 JN897589 JN897590 JN897591 JN897592 JN897593 AET96245.1 JN897606 JN897607 JN897610 AET96259.1 JN897545 JN897615 JN897617 JN897618 AET96271.1 JN897542 JN897544 AET96197.1 JN897599 JN897601 JN897602 AET96254.1 JN897605 JN897608 JN897609 AET96258.1 JN897548 JN897549 JN897550 JN897551 AET96202.1 JN897604 AET96257.1 JN898022 AET96675.1 JN898015 AET96668.1 JN898038 AET96691.1 JN898020 AET96673.1 JN898009 JN898014 AET96667.1 JN898023 AET96676.1 JN898008 AET96661.1 JN898030 JN898032 JN898033 JN898034 AET96685.1 JN898016 JN898018 AET96669.1 JN898045 JN898046 JN898047 JN898048 AET96698.1 JN898043 JN898044 JN898050 JN898051 JN898052 JN898053 JN898054 AET96707.1 JN898055 JN898056 JN898057 JN898058 JN898059 AET96711.1 JN898025 JN898026 JN898027 JN898028 JN898029 AET96681.1 JN898011 JN898013 JN898060 JN898061 JN898062 JN898063 AET96716.1 JN898006 JN898007 AET96659.1 JN897541 AET96194.1 JN897547 AET96200.1 JN898037 AET96690.1 JN898021 AET96674.1 JN898049 AET96702.1 JN897598 AET96251.1 JN898024 AET96677.1 JN897546 AET96199.1 JN897565 AET96218.1 JN897584 AET96237.1 JN897561 AET96214.1 JN898031 AET96684.1 JN897587 AET96240.1 JN897570 AET96223.1 JN897566 AET96219.1 JN898042 AET96695.1 JN898036 AET96689.1 JN897572 AET96225.1 JN898004 AET96657.1 AK417638 BAN20853.1 GDKW01000104 JAI56491.1 GECL01003165 JAP02959.1 GAHY01000504 JAA77006.1 GFTR01005673 JAW10753.1 GEDC01021813 JAS15485.1 KE164279 EPQ16653.1 AAPE02030108 AKHW03006582 KYO19679.1 KB031037 ELK06049.1 GAAZ01001934 JAA96009.1
Proteomes
PRIDE
SUPFAM
SSF53300
SSF53300
Gene 3D
ProteinModelPortal
Q2F5X3
D0AB76
L7X3X7
A0A212FKP4
A0A2A4JIA6
A0A2H1X328
+ More
S4P8H2 A0A194QXK8 A0A194QAX9 E3UKM5 A0A0L7LIK5 D8VIK0 D8VIM0 D8VII0 D8VIK9 D8VIJ9 D8VII1 D8VIK1 G9JQN6 G9JPI5 G9JQQ4 G9JQN1 G9JQT3 G9JQN8 G9JQQ9 G9JQM2 G9JPG9 G9JPE6 G9JPH2 G9JPE0 G9JPG6 G9JPF4 G9JPD8 G9JPE7 G9JPF5 G9JPC3 G9JPC8 G9JPG1 G9JPH5 G9JPI7 G9JPB3 G9JPH0 G9JPH4 G9JPB8 G9JPH3 G9JQP1 G9JQN4 G9JQQ7 G9JQN9 G9JQN3 G9JQP2 G9JQM7 G9JQQ1 G9JQN5 G9JQR4 G9JQS3 G9JQS7 G9JQP7 G9JQT2 G9JQM5 G9JPB0 G9JPB6 G9JQQ6 G9JQP0 G9JQR8 G9JPG7 G9JQP3 G9JPB5 G9JPD4 G9JPF3 G9JPD0 G9JQQ0 G9JPF6 G9JPD9 G9JPD5 G9JQR1 G9JQQ5 G9JPE1 G9JQM3 R4WDF5 A0A0P4VUT1 A0A0V0G6L2 R4FQB0 A0A224XRH9 A0A1B6CQ59 A0A1S3H8E6 A0A1S3H700 S7Q067 G1PRQ7 A0A151M585 L5K2W8 T1D9J7
S4P8H2 A0A194QXK8 A0A194QAX9 E3UKM5 A0A0L7LIK5 D8VIK0 D8VIM0 D8VII0 D8VIK9 D8VIJ9 D8VII1 D8VIK1 G9JQN6 G9JPI5 G9JQQ4 G9JQN1 G9JQT3 G9JQN8 G9JQQ9 G9JQM2 G9JPG9 G9JPE6 G9JPH2 G9JPE0 G9JPG6 G9JPF4 G9JPD8 G9JPE7 G9JPF5 G9JPC3 G9JPC8 G9JPG1 G9JPH5 G9JPI7 G9JPB3 G9JPH0 G9JPH4 G9JPB8 G9JPH3 G9JQP1 G9JQN4 G9JQQ7 G9JQN9 G9JQN3 G9JQP2 G9JQM7 G9JQQ1 G9JQN5 G9JQR4 G9JQS3 G9JQS7 G9JQP7 G9JQT2 G9JQM5 G9JPB0 G9JPB6 G9JQQ6 G9JQP0 G9JQR8 G9JPG7 G9JQP3 G9JPB5 G9JPD4 G9JPF3 G9JPD0 G9JQQ0 G9JPF6 G9JPD9 G9JPD5 G9JQR1 G9JQQ5 G9JPE1 G9JQM3 R4WDF5 A0A0P4VUT1 A0A0V0G6L2 R4FQB0 A0A224XRH9 A0A1B6CQ59 A0A1S3H8E6 A0A1S3H700 S7Q067 G1PRQ7 A0A151M585 L5K2W8 T1D9J7
PDB
6EPF
E-value=3.41207e-97,
Score=906
Ontologies
KEGG
Topology
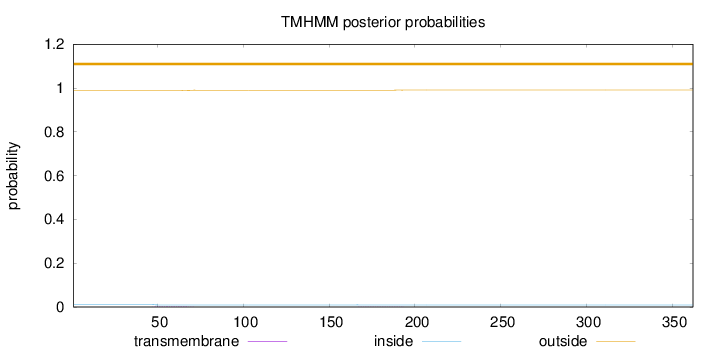
Length:
362
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05998
Exp number, first 60 AAs:
0.0268
Total prob of N-in:
0.01166
outside
1 - 362
Population Genetic Test Statistics
Pi
234.048557
Theta
220.914202
Tajima's D
0.25992
CLR
0.410686
CSRT
0.440077996100195
Interpretation
Uncertain
