Gene
KWMTBOMO10128 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005648
Annotation
Similar_to_CG4692_[Heliconius_melpomene]
Full name
Putative ATP synthase subunit f, mitochondrial
Location in the cell
Mitochondrial Reliability : 2.408
Sequence
CDS
ATGGCTTTCGGTGATTACCCCAAGGAGTATAATCCTGCTGTACATGGGCCTTACGATCCTGCACGCTACTATGGAAAACCTGATACTCCATTCAGTCAGTTGAAGCTGAACGAAATCGGTTCGTGGTTCGGTCGCCGCAGCAAGACTCCGTCTGCCGTGGCCGGAGCTTTCAGTCGAGCCTGGTGGAGATGGCAACATAAGTACGTACAACCTAAGAAGGTTGGCATGGCTCCATTCTATCAACTACTCGTTGGCAGCATGGTCTTCTTCTATGCCATCAACTATGGAAGAATTAAGCACCACAAGAACTACAAATACCACTAA
Protein
MAFGDYPKEYNPAVHGPYDPARYYGKPDTPFSQLKLNEIGSWFGRRSKTPSAVAGAFSRAWWRWQHKYVQPKKVGMAPFYQLLVGSMVFFYAINYGRIKHHKNYKYH
Summary
Description
Mitochondrial membrane ATP synthase (F(1)F(0) ATP synthase or Complex V) produces ATP from ADP in the presence of a proton gradient across the membrane which is generated by electron transport complexes of the respiratory chain. F-type ATPases consist of two structural domains, F(1) - containing the extramembraneous catalytic core and F(0) - containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. During catalysis, ATP synthesis in the catalytic domain of F(1) is coupled via a rotary mechanism of the central stalk subunits to proton translocation. Part of the complex F(0) domain. Minor subunit located with subunit a in the membrane (By similarity).
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(0) seems to have nine subunits: a, b, c, d, e, f, g, F6 and 8 (or A6L) (By similarity).
Similarity
Belongs to the ATPase F chain family.
Keywords
Alternative splicing
ATP synthesis
Complete proteome
Hydrogen ion transport
Ion transport
Membrane
Mitochondrion
Reference proteome
Transport
Feature
chain Putative ATP synthase subunit f, mitochondrial
splice variant In isoform B.
splice variant In isoform B.
Uniprot
L7X0S3
D0AB75
I4DK72
H9J806
A0A194QS47
A0A2H1X381
+ More
A0A2A4JHT3 A0A1E1WAH0 A0A194QGP8 U5EZG0 A0A182YPR7 T1GFI1 A0A182MB01 A0A182QWK6 T1PHQ3 A0A0K8TSU5 A0A182RJV0 A0A182WAY1 A0A1A9VU72 A0A1B0AF80 A0A182SSR1 A0A1L8E0A7 Q1HRJ7 A0A182PUS2 A0A1A9XPT4 A0A1B0BRL6 A0A1A9X4V4 A0A067R7N7 A0A336JVX2 B4LJI2 V5GXI9 C6ZQT5 A0A1L8EFK9 A0A182JLI5 A0A212FKP6 A0A2M4A5B9 A0A2M4C557 W5JQR1 B8RJ20 A0A3B0JRI7 T1D5H4 A0A182IGF7 A0A2M3Z0R8 B4MP00 B5RJ63 A0A1W4WCK4 Q6XI17 Q9W141 W8CDF0 A0A1B0G437 A0A0L0C0Z6 B4J809 B0WDF3 B4KQ55 B4H565 T1DHH4 B4IH47 B4QCG5 Q28X92 A0A1I8M533 A0A182XT52 Q7QLC5 A0A0C9RRE6 B3NQV2 B3MG91 A0A023ECZ6 A0A034WWD1 A0A1I8PV83 A0A0A1XR83 A0A0A1WMS9 A0A0K8W557 A0A293MUV9 A0A1J1IH77 A0A182NQ92 Q9W141-2 A0A1B0CFL2 A0A2J7RIS2 A0A2M4B0E3 A0A2R5LEX6 A0A0K8ULD5 A0A2H8TX46 A0A0T6B3W9 B5U263 A0A195EW94 R4G5B0 B5U240 B5U249 A0A0A9YI81 A0A146LF38 B5U218 B5U217 B5U235 A0A2I9LNS8 Q201W3 A0A1W4WV18 J3JV39 A0A151X1X8 Q09JM8 D6WX58 R4WQJ6
A0A2A4JHT3 A0A1E1WAH0 A0A194QGP8 U5EZG0 A0A182YPR7 T1GFI1 A0A182MB01 A0A182QWK6 T1PHQ3 A0A0K8TSU5 A0A182RJV0 A0A182WAY1 A0A1A9VU72 A0A1B0AF80 A0A182SSR1 A0A1L8E0A7 Q1HRJ7 A0A182PUS2 A0A1A9XPT4 A0A1B0BRL6 A0A1A9X4V4 A0A067R7N7 A0A336JVX2 B4LJI2 V5GXI9 C6ZQT5 A0A1L8EFK9 A0A182JLI5 A0A212FKP6 A0A2M4A5B9 A0A2M4C557 W5JQR1 B8RJ20 A0A3B0JRI7 T1D5H4 A0A182IGF7 A0A2M3Z0R8 B4MP00 B5RJ63 A0A1W4WCK4 Q6XI17 Q9W141 W8CDF0 A0A1B0G437 A0A0L0C0Z6 B4J809 B0WDF3 B4KQ55 B4H565 T1DHH4 B4IH47 B4QCG5 Q28X92 A0A1I8M533 A0A182XT52 Q7QLC5 A0A0C9RRE6 B3NQV2 B3MG91 A0A023ECZ6 A0A034WWD1 A0A1I8PV83 A0A0A1XR83 A0A0A1WMS9 A0A0K8W557 A0A293MUV9 A0A1J1IH77 A0A182NQ92 Q9W141-2 A0A1B0CFL2 A0A2J7RIS2 A0A2M4B0E3 A0A2R5LEX6 A0A0K8ULD5 A0A2H8TX46 A0A0T6B3W9 B5U263 A0A195EW94 R4G5B0 B5U240 B5U249 A0A0A9YI81 A0A146LF38 B5U218 B5U217 B5U235 A0A2I9LNS8 Q201W3 A0A1W4WV18 J3JV39 A0A151X1X8 Q09JM8 D6WX58 R4WQJ6
Pubmed
23674305
22651552
19121390
26354079
25244985
26369729
+ More
17204158 24845553 17994087 20496585 22118469 20920257 23761445 24330624 14525923 17550304 10731132 12537572 12537569 24495485 26108605 18057021 22936249 15632085 25315136 9087549 12364791 14747013 17210077 24945155 26483478 25348373 25830018 20087394 25401762 26823975 20075255 29248469 22516182 23537049 18070664 18362917 19820115 23691247
17204158 24845553 17994087 20496585 22118469 20920257 23761445 24330624 14525923 17550304 10731132 12537572 12537569 24495485 26108605 18057021 22936249 15632085 25315136 9087549 12364791 14747013 17210077 24945155 26483478 25348373 25830018 20087394 25401762 26823975 20075255 29248469 22516182 23537049 18070664 18362917 19820115 23691247
EMBL
KC469893
AGC92709.1
FP102339
CBH09270.1
AK401690
BAM18312.1
+ More
BABH01020219 KQ461155 KPJ08312.1 ODYU01013053 SOQ59708.1 NWSH01001365 PCG71531.1 GDQN01010646 GDQN01007054 JAT80408.1 JAT84000.1 KQ459232 KPJ02621.1 GANO01000864 JAB59007.1 CAQQ02393081 AXCM01004513 AXCN02001889 KA647418 AFP62047.1 GDAI01000171 JAI17432.1 GFDF01001977 JAV12107.1 DQ440097 ABF18130.1 JXJN01019147 KK852652 KDR19410.1 UFQS01000003 UFQT01000003 SSW96774.1 SSX17161.1 CH940648 EDW61550.1 GALX01001979 JAB66487.1 EZ114886 ACU30939.1 GFDG01001456 JAV17343.1 AXCP01007179 AGBW02008005 OWR54314.1 GGFK01002620 MBW35941.1 GGFJ01010977 MBW60118.1 ADMH02000608 ETN65643.1 EZ000445 ACJ64319.1 OUUW01000001 SPP73748.1 GALA01000508 JAA94344.1 APCN01008223 GGFM01001354 MBW22105.1 CH963848 EDW73839.1 BT044337 ACH92402.1 AY232015 CM000158 AAR10038.1 EDW92569.1 AE013599 AY060738 BT025808 BT030456 GAMC01002061 JAC04495.1 CCAG010002612 JRES01001049 KNC25965.1 CH916367 EDW02239.1 DS231897 EDS44524.1 CH933808 EDW09183.2 KRG04509.1 CH479210 EDW32901.1 GAMD01002341 JAA99249.1 CH480838 EDW49223.1 CM000362 CM002911 EDX08581.1 KMY96383.1 CM000071 EAL26424.1 AAAB01007639 EAA03027.1 GBYB01009906 JAG79673.1 CH954179 EDV57035.1 CH902619 EDV37794.1 JXUM01149425 GAPW01006341 GAPW01006340 KQ570772 JAC07257.1 KXJ68304.1 GAKP01000844 JAC58108.1 GBXI01000815 JAD13477.1 GBXI01014559 JAC99732.1 GDHF01012957 GDHF01006088 JAI39357.1 JAI46226.1 GFWV01019895 MAA44623.1 CVRI01000047 CRK98390.1 AJWK01010169 AJWK01010170 NEVH01003493 PNF40738.1 GGFK01013121 MBW46442.1 GGLE01003958 MBY08084.1 GDHF01024807 JAI27507.1 GFXV01006694 MBW18499.1 LJIG01009932 KRT82092.1 FJ158240 FJ158241 FJ158242 FJ158243 FJ158244 FJ158245 GQ336322 GQ336323 GQ336324 GQ336325 GQ336326 ACH99579.1 ACH99580.1 ACH99581.1 ACH99582.1 ACH99583.1 ACH99584.1 ACU11636.1 ACU11637.1 ACU11638.1 ACU11639.1 ACU11640.1 KQ981954 KYN32162.1 GAHY01000101 JAA77409.1 FJ158217 FJ158221 FJ158223 GQ336320 GQ336321 ACH99556.1 ACH99560.1 ACH99562.1 ACU11634.1 ACU11635.1 FJ158226 FJ158227 FJ158228 FJ158229 FJ158230 FJ158231 FJ158232 FJ158233 FJ158234 FJ158235 FJ158236 FJ158237 FJ158238 FJ158239 ACH99565.1 ACH99566.1 ACH99567.1 ACH99568.1 ACH99569.1 ACH99570.1 ACH99571.1 ACH99572.1 ACH99573.1 ACH99574.1 ACH99575.1 ACH99576.1 ACH99577.1 ACH99578.1 GBHO01010832 JAG32772.1 GDHC01013027 JAQ05602.1 FJ158195 FJ158202 FJ158204 FJ158208 FJ158209 FJ158210 FJ158211 GU201928 ACH99534.1 ACH99541.1 ACH99543.1 ACH99547.1 ACH99548.1 ACH99549.1 ACH99550.1 ACZ95802.1 FJ158194 FJ158196 FJ158197 FJ158198 FJ158199 FJ158200 FJ158201 FJ158203 FJ158205 FJ158206 FJ158207 ACH99533.1 ACH99535.1 ACH99536.1 ACH99537.1 ACH99538.1 ACH99539.1 ACH99540.1 ACH99542.1 ACH99544.1 ACH99545.1 ACH99546.1 FJ158212 FJ158213 FJ158214 FJ158215 FJ158216 FJ158218 FJ158219 FJ158220 FJ158222 FJ158224 FJ158225 ACH99551.1 ACH99552.1 ACH99553.1 ACH99554.1 ACH99555.1 ACH99557.1 ACH99558.1 ACH99559.1 ACH99561.1 ACH99563.1 ACH99564.1 GFWZ01000051 MBW20041.1 ABLF02027176 DQ413223 AK340420 ABD72691.1 BAH71034.1 APGK01044595 BT127105 KB741027 KB632186 AEE62067.1 ENN74885.1 ERL89730.1 KQ982585 KYQ54194.1 DQ886818 ABI52735.1 KQ971361 EFA09167.1 AK416962 BAN20177.1
BABH01020219 KQ461155 KPJ08312.1 ODYU01013053 SOQ59708.1 NWSH01001365 PCG71531.1 GDQN01010646 GDQN01007054 JAT80408.1 JAT84000.1 KQ459232 KPJ02621.1 GANO01000864 JAB59007.1 CAQQ02393081 AXCM01004513 AXCN02001889 KA647418 AFP62047.1 GDAI01000171 JAI17432.1 GFDF01001977 JAV12107.1 DQ440097 ABF18130.1 JXJN01019147 KK852652 KDR19410.1 UFQS01000003 UFQT01000003 SSW96774.1 SSX17161.1 CH940648 EDW61550.1 GALX01001979 JAB66487.1 EZ114886 ACU30939.1 GFDG01001456 JAV17343.1 AXCP01007179 AGBW02008005 OWR54314.1 GGFK01002620 MBW35941.1 GGFJ01010977 MBW60118.1 ADMH02000608 ETN65643.1 EZ000445 ACJ64319.1 OUUW01000001 SPP73748.1 GALA01000508 JAA94344.1 APCN01008223 GGFM01001354 MBW22105.1 CH963848 EDW73839.1 BT044337 ACH92402.1 AY232015 CM000158 AAR10038.1 EDW92569.1 AE013599 AY060738 BT025808 BT030456 GAMC01002061 JAC04495.1 CCAG010002612 JRES01001049 KNC25965.1 CH916367 EDW02239.1 DS231897 EDS44524.1 CH933808 EDW09183.2 KRG04509.1 CH479210 EDW32901.1 GAMD01002341 JAA99249.1 CH480838 EDW49223.1 CM000362 CM002911 EDX08581.1 KMY96383.1 CM000071 EAL26424.1 AAAB01007639 EAA03027.1 GBYB01009906 JAG79673.1 CH954179 EDV57035.1 CH902619 EDV37794.1 JXUM01149425 GAPW01006341 GAPW01006340 KQ570772 JAC07257.1 KXJ68304.1 GAKP01000844 JAC58108.1 GBXI01000815 JAD13477.1 GBXI01014559 JAC99732.1 GDHF01012957 GDHF01006088 JAI39357.1 JAI46226.1 GFWV01019895 MAA44623.1 CVRI01000047 CRK98390.1 AJWK01010169 AJWK01010170 NEVH01003493 PNF40738.1 GGFK01013121 MBW46442.1 GGLE01003958 MBY08084.1 GDHF01024807 JAI27507.1 GFXV01006694 MBW18499.1 LJIG01009932 KRT82092.1 FJ158240 FJ158241 FJ158242 FJ158243 FJ158244 FJ158245 GQ336322 GQ336323 GQ336324 GQ336325 GQ336326 ACH99579.1 ACH99580.1 ACH99581.1 ACH99582.1 ACH99583.1 ACH99584.1 ACU11636.1 ACU11637.1 ACU11638.1 ACU11639.1 ACU11640.1 KQ981954 KYN32162.1 GAHY01000101 JAA77409.1 FJ158217 FJ158221 FJ158223 GQ336320 GQ336321 ACH99556.1 ACH99560.1 ACH99562.1 ACU11634.1 ACU11635.1 FJ158226 FJ158227 FJ158228 FJ158229 FJ158230 FJ158231 FJ158232 FJ158233 FJ158234 FJ158235 FJ158236 FJ158237 FJ158238 FJ158239 ACH99565.1 ACH99566.1 ACH99567.1 ACH99568.1 ACH99569.1 ACH99570.1 ACH99571.1 ACH99572.1 ACH99573.1 ACH99574.1 ACH99575.1 ACH99576.1 ACH99577.1 ACH99578.1 GBHO01010832 JAG32772.1 GDHC01013027 JAQ05602.1 FJ158195 FJ158202 FJ158204 FJ158208 FJ158209 FJ158210 FJ158211 GU201928 ACH99534.1 ACH99541.1 ACH99543.1 ACH99547.1 ACH99548.1 ACH99549.1 ACH99550.1 ACZ95802.1 FJ158194 FJ158196 FJ158197 FJ158198 FJ158199 FJ158200 FJ158201 FJ158203 FJ158205 FJ158206 FJ158207 ACH99533.1 ACH99535.1 ACH99536.1 ACH99537.1 ACH99538.1 ACH99539.1 ACH99540.1 ACH99542.1 ACH99544.1 ACH99545.1 ACH99546.1 FJ158212 FJ158213 FJ158214 FJ158215 FJ158216 FJ158218 FJ158219 FJ158220 FJ158222 FJ158224 FJ158225 ACH99551.1 ACH99552.1 ACH99553.1 ACH99554.1 ACH99555.1 ACH99557.1 ACH99558.1 ACH99559.1 ACH99561.1 ACH99563.1 ACH99564.1 GFWZ01000051 MBW20041.1 ABLF02027176 DQ413223 AK340420 ABD72691.1 BAH71034.1 APGK01044595 BT127105 KB741027 KB632186 AEE62067.1 ENN74885.1 ERL89730.1 KQ982585 KYQ54194.1 DQ886818 ABI52735.1 KQ971361 EFA09167.1 AK416962 BAN20177.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000076408
UP000015102
+ More
UP000075883 UP000075886 UP000075900 UP000075920 UP000078200 UP000092445 UP000075901 UP000075885 UP000092443 UP000092460 UP000091820 UP000027135 UP000008792 UP000075880 UP000007151 UP000000673 UP000268350 UP000075840 UP000007798 UP000192221 UP000002282 UP000000803 UP000092444 UP000037069 UP000001070 UP000002320 UP000009192 UP000008744 UP000001292 UP000000304 UP000001819 UP000095301 UP000076407 UP000007062 UP000008711 UP000007801 UP000069940 UP000249989 UP000095300 UP000183832 UP000075884 UP000092461 UP000235965 UP000078541 UP000002358 UP000007819 UP000192223 UP000019118 UP000030742 UP000075809 UP000007266
UP000075883 UP000075886 UP000075900 UP000075920 UP000078200 UP000092445 UP000075901 UP000075885 UP000092443 UP000092460 UP000091820 UP000027135 UP000008792 UP000075880 UP000007151 UP000000673 UP000268350 UP000075840 UP000007798 UP000192221 UP000002282 UP000000803 UP000092444 UP000037069 UP000001070 UP000002320 UP000009192 UP000008744 UP000001292 UP000000304 UP000001819 UP000095301 UP000076407 UP000007062 UP000008711 UP000007801 UP000069940 UP000249989 UP000095300 UP000183832 UP000075884 UP000092461 UP000235965 UP000078541 UP000002358 UP000007819 UP000192223 UP000019118 UP000030742 UP000075809 UP000007266
Interpro
Gene 3D
CDD
ProteinModelPortal
L7X0S3
D0AB75
I4DK72
H9J806
A0A194QS47
A0A2H1X381
+ More
A0A2A4JHT3 A0A1E1WAH0 A0A194QGP8 U5EZG0 A0A182YPR7 T1GFI1 A0A182MB01 A0A182QWK6 T1PHQ3 A0A0K8TSU5 A0A182RJV0 A0A182WAY1 A0A1A9VU72 A0A1B0AF80 A0A182SSR1 A0A1L8E0A7 Q1HRJ7 A0A182PUS2 A0A1A9XPT4 A0A1B0BRL6 A0A1A9X4V4 A0A067R7N7 A0A336JVX2 B4LJI2 V5GXI9 C6ZQT5 A0A1L8EFK9 A0A182JLI5 A0A212FKP6 A0A2M4A5B9 A0A2M4C557 W5JQR1 B8RJ20 A0A3B0JRI7 T1D5H4 A0A182IGF7 A0A2M3Z0R8 B4MP00 B5RJ63 A0A1W4WCK4 Q6XI17 Q9W141 W8CDF0 A0A1B0G437 A0A0L0C0Z6 B4J809 B0WDF3 B4KQ55 B4H565 T1DHH4 B4IH47 B4QCG5 Q28X92 A0A1I8M533 A0A182XT52 Q7QLC5 A0A0C9RRE6 B3NQV2 B3MG91 A0A023ECZ6 A0A034WWD1 A0A1I8PV83 A0A0A1XR83 A0A0A1WMS9 A0A0K8W557 A0A293MUV9 A0A1J1IH77 A0A182NQ92 Q9W141-2 A0A1B0CFL2 A0A2J7RIS2 A0A2M4B0E3 A0A2R5LEX6 A0A0K8ULD5 A0A2H8TX46 A0A0T6B3W9 B5U263 A0A195EW94 R4G5B0 B5U240 B5U249 A0A0A9YI81 A0A146LF38 B5U218 B5U217 B5U235 A0A2I9LNS8 Q201W3 A0A1W4WV18 J3JV39 A0A151X1X8 Q09JM8 D6WX58 R4WQJ6
A0A2A4JHT3 A0A1E1WAH0 A0A194QGP8 U5EZG0 A0A182YPR7 T1GFI1 A0A182MB01 A0A182QWK6 T1PHQ3 A0A0K8TSU5 A0A182RJV0 A0A182WAY1 A0A1A9VU72 A0A1B0AF80 A0A182SSR1 A0A1L8E0A7 Q1HRJ7 A0A182PUS2 A0A1A9XPT4 A0A1B0BRL6 A0A1A9X4V4 A0A067R7N7 A0A336JVX2 B4LJI2 V5GXI9 C6ZQT5 A0A1L8EFK9 A0A182JLI5 A0A212FKP6 A0A2M4A5B9 A0A2M4C557 W5JQR1 B8RJ20 A0A3B0JRI7 T1D5H4 A0A182IGF7 A0A2M3Z0R8 B4MP00 B5RJ63 A0A1W4WCK4 Q6XI17 Q9W141 W8CDF0 A0A1B0G437 A0A0L0C0Z6 B4J809 B0WDF3 B4KQ55 B4H565 T1DHH4 B4IH47 B4QCG5 Q28X92 A0A1I8M533 A0A182XT52 Q7QLC5 A0A0C9RRE6 B3NQV2 B3MG91 A0A023ECZ6 A0A034WWD1 A0A1I8PV83 A0A0A1XR83 A0A0A1WMS9 A0A0K8W557 A0A293MUV9 A0A1J1IH77 A0A182NQ92 Q9W141-2 A0A1B0CFL2 A0A2J7RIS2 A0A2M4B0E3 A0A2R5LEX6 A0A0K8ULD5 A0A2H8TX46 A0A0T6B3W9 B5U263 A0A195EW94 R4G5B0 B5U240 B5U249 A0A0A9YI81 A0A146LF38 B5U218 B5U217 B5U235 A0A2I9LNS8 Q201W3 A0A1W4WV18 J3JV39 A0A151X1X8 Q09JM8 D6WX58 R4WQJ6
Ontologies
PATHWAY
PANTHER
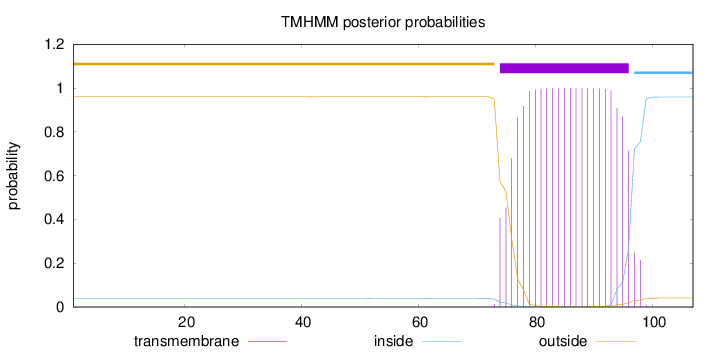
Topology
Subcellular location
Mitochondrion membrane
Length:
107
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.25672
Exp number, first 60 AAs:
0.00871
Total prob of N-in:
0.04010
outside
1 - 73
TMhelix
74 - 96
inside
97 - 107
Population Genetic Test Statistics
Pi
186.168416
Theta
155.0938
Tajima's D
-0.6321
CLR
0.137563
CSRT
0.214339283035848
Interpretation
Uncertain
