Gene
KWMTBOMO10125
Pre Gene Modal
BGIBMGA005649
Annotation
PREDICTED:_dynein_assembly_factor_5?_axonemal_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.644 Golgi Reliability : 1.276
Sequence
CDS
ATGTCTAAAACTAAATGTCCTTTGGAAGAAAATGCTCGACGTGTTCTTGAACAATATATAACGGCTTTACAAAGCGAATCAAAGATAGCGCGAAAAAATGCGCTCACGAAGTTAAGCGAAGAAATTTTTGAGAATCCTGAAAATGTTGACTGTGATCTCACTGTGGTGTTTCCAGAAATCTACGCGCACGTACTCAAAAGCTATTCCGATCAGTCGGAGGCGTGTCGAGAATTAGCCGTGCAAATAATTTCGAACTTCATAGAAAAGCTCCCACTGAATGACTATTATTTAACCTATATATTTCCTACGCTTGTCCGACGTATCGGATGTCCTGAGATTGTGGAAGAATCTGAAGAGATTCGGCTAGTTTTAATCCAGCTAGTTCACACTATCTTAGATAGATACAAAGTTACACATTACTTAAATTCTTTTCTAAATGATTTAACAAGTATTTTAACCAAAACGGTTACGGATCCTTATTCTAAAGTGAAGGTAGAAGCATGTGAATGCATTATATTGTTAGCTGAGAACTTGCAGCGGGATTTTCATTTTCAATCTGAGAGCTATGTGAAGCCCGTGCTGTCAAATCTAGCACACCAGCATTTTAGGGTTCGGGCTGCAGCTGTTAAAGCTATAGGAGCAATAGTGATGTCAGGAAATGTGAAATGCTTAGAGCTATCAATAACACCAATGGCAGAGAAGCTGTTTGATGAAAATAGTCAAGTTCGCCTCCAGGTGACATTAGAAGTAGGAAACTGGATGTTAAAATACAGAGACCGATATTCTTTTTGGCATCGCATGATGCCTCTTTTGTTAACTAGCCTGAGTGACGTAATGGCTAGCATCAGGGAAACTGCCACGAAACTCTGGGATGATGTTGGCTTACAATACATGGAGGAAAATGAGGAAGATTTAAAAAAGAAGAGTGATTTCCTGAAAGATGTTCCCTCGCATTATCCTGATGTTAAAAGACCCAACATGGGTTGTAGAGTATTGGTGCAGAGTAATATTGGGAAGATTGTTCCTGCTGTTGAAAAAGAAATGGAGGGCTGGCAAGCAGATGCTCGTCTCCGCGTGGCTCAGCTCCTCTGCTGGTTGATATTATGCGCCGAGGAGGGTTCTACACAGCATGCCAACCCTATTGTTCGTACTATGATACGTGGGGCAGCTGATGAAGATCCTAGAGTTGTGGTCGAGGTGAAACGCGCTGCAGAATTATTCGGATACTTCATCACTCCGGAAACGTGGTGGCCGCTGCTGGAGGCCGACGTGGAATCGTGGGCAGTCTTGCTTGTCCTAGCCAACATCATCAAAGGTTCTCGTGTGGAACTTGTGAAAGGGAAGATTTTGACTGAGATATGTAAAGAACTGGCGGATCCTGACCGTTGTCGGACTAGAAAGCCAAAGTACCAGACGAATTTGCTTCACGTATCGGAGGCGTTGATGGAGCTCTGCGGTGACGAGTGTGGGTCAGTCGCCAACGATCTTTTCACCATCAACTTCACTGTTTACGCCATGCCCGTCGATGATCAGATACAGTTTATGGCGTTGTCAAACCTGGACAGACTTCGGTGCATTGAAAAGTCCGGTAAAACATTAACCAGTCTCTATGAAAGACACATCGGTAAAGTGCTCGCCAACATCACCAGCGACGCCCCCACGTGGACTCTGCTCACCCCCGATCGGTGTCTGCTTGAATGCGTTCTGATGCACGCAGGTTCAGCCATGGGAGCCCAGCTGCACCTCATAGCTCCCCTATTAAAGGAGTGTCTGGCGAACACGGAAACGGACGCGGAAGTCAGACTAAAGATATTCACGACTCTATCGACTGCTCTGCTGAAGAGACAGGAGAACTTCAGGAAATGCGACAGTGACAAACTTGGGGCCTTCCTGAAGATCATCATAGAAGAAATAATAATGCCTAACATGATATGGTCGGCGGGTCGCACCGCTGAGGCGATACGCACCGCGGCCGTCGCGTGTCTGTGCTCCGCGCTGCAGGACAGCCCTCAGGACGAGCGCATCCGGGCCGAGAACGGTGGAGATGGCGACACCGTCGATAAGGAAGTAAACCTTTTTCCCACAAAAGAATCCTTGGAACCGTTTTTGGACGAAATGGTGCCGCTCCTCATGGGTCTGGTGGAGGACAACTCGACGTTGACGAGACAGCACACTCTGCGTGCCGTGTGCTGCCTCGCGACTTTAGCCCGCCAACGCGGCTGCTTTACAGCGGATATATTGAATAAAATGTATTTCGTTGTATTGAAACGCCTCGACGACGTTAGCGACAAAGTTCGTTCTTACGCCGTGCAGACTTTATGTAATTTGTTCAAGAACCGACCGCAGCCGTACGATACGACTGTATACGGTGCGCACGTTGACGCTCTGTACTCCGCCATGTTGGTGCACCTGGATGACGTCGATGAAACGTTCAGAAATGAAATGTTAGATGCTTTACTCCAGCTCAGCGATATCGATCCAAGAACGTTAATGAAGAAAGTGAAATCGAATATTCACGTTTATCGAAATAAATCCGCTTACGAGAAGATATCGAGTCACCTTGAGAAGCTGACCATATGA
Protein
MSKTKCPLEENARRVLEQYITALQSESKIARKNALTKLSEEIFENPENVDCDLTVVFPEIYAHVLKSYSDQSEACRELAVQIISNFIEKLPLNDYYLTYIFPTLVRRIGCPEIVEESEEIRLVLIQLVHTILDRYKVTHYLNSFLNDLTSILTKTVTDPYSKVKVEACECIILLAENLQRDFHFQSESYVKPVLSNLAHQHFRVRAAAVKAIGAIVMSGNVKCLELSITPMAEKLFDENSQVRLQVTLEVGNWMLKYRDRYSFWHRMMPLLLTSLSDVMASIRETATKLWDDVGLQYMEENEEDLKKKSDFLKDVPSHYPDVKRPNMGCRVLVQSNIGKIVPAVEKEMEGWQADARLRVAQLLCWLILCAEEGSTQHANPIVRTMIRGAADEDPRVVVEVKRAAELFGYFITPETWWPLLEADVESWAVLLVLANIIKGSRVELVKGKILTEICKELADPDRCRTRKPKYQTNLLHVSEALMELCGDECGSVANDLFTINFTVYAMPVDDQIQFMALSNLDRLRCIEKSGKTLTSLYERHIGKVLANITSDAPTWTLLTPDRCLLECVLMHAGSAMGAQLHLIAPLLKECLANTETDAEVRLKIFTTLSTALLKRQENFRKCDSDKLGAFLKIIIEEIIMPNMIWSAGRTAEAIRTAAVACLCSALQDSPQDERIRAENGGDGDTVDKEVNLFPTKESLEPFLDEMVPLLMGLVEDNSTLTRQHTLRAVCCLATLARQRGCFTADILNKMYFVVLKRLDDVSDKVRSYAVQTLCNLFKNRPQPYDTTVYGAHVDALYSAMLVHLDDVDETFRNEMLDALLQLSDIDPRTLMKKVKSNIHVYRNKSAYEKISSHLEKLTI
Summary
Uniprot
A0A173FDM6
E2GIU6
A0A2A4JIY0
D0AB73
L7WWA6
H9J807
+ More
A0A212FKQ7 A0A194QB14 A0A2H1X389 A0A2J7RIS0 A0A067R4R1 A0A1Y1LFQ5 A0A2P8XLI2 A0A026WLP7 E2AAI4 F4WFK7 A0A1Y1LJH1 A0A3L8DXE8 D6WZ28 A0A158NSI2 A0A195CTV0 A0A151J434 A0A195FUE5 K7IMZ7 A0A232ELJ6 A0A151WIL1 A0A151HYP6 E0VSR2 A0A1S3J8R9 A0A1S3K505 E2BW17 A0A2A3E6E3 A0A2C9JQQ3 V3Z8D9 W4XCH6 C3Y8Q6 K1P6F0 R7UI93 A0A0C9PZJ6 F6S428 A0A1L8EQB6 W5M0E5 A0A1L8EY17 A0A0L7R775 A0JMW2 A0A1B6GW16 A0A2B4RBP0 A0A0L8G7D7 H2S9A9 A0A210QL21 A0A3P9B6Z3 A0A3P8RJF3 A0A3B3RXV3 A0A3Q2WTL2 I3JCA3 N6TFE4 U4U796 A0A2T7PY40 A0A3M6TCZ2 H3B2F2 W5LCW1 A0A3B3WNP8 A0A2G8KC10 A0A3B4EUY4 A0A087Y2B7 H2YGR6 W5UCV2 A0A3B4DSU1 A0A146RJF3 A0A147AU06 A0A3Q3GPL0 M3ZQM0 A0A3B3TZJ0 A0A146RM00 A0A146QY71 A0A3B3RXT1 A0A3B5AZ10 A0A3Q4GCM0 A0A1S3S1P0 F6Q2F8 A0A3Q1GP08 A0A3Q0S811 A0A1W2WEK6 A0A0F8CDB1 A0A3B5REA6 A0A096M6B0 A0A3B4U7X3 A0A0F8CYL3 A0A3Q1JUX1 A0A1B6CAU1 A0A3P8SES1 A0A3Q2XHN2 A0A1A8I9T1 A0A1A8N110 A0A1A8UI62 A0A1A7ZL32 A0A315WDI0 A0A1A8ERE0
A0A212FKQ7 A0A194QB14 A0A2H1X389 A0A2J7RIS0 A0A067R4R1 A0A1Y1LFQ5 A0A2P8XLI2 A0A026WLP7 E2AAI4 F4WFK7 A0A1Y1LJH1 A0A3L8DXE8 D6WZ28 A0A158NSI2 A0A195CTV0 A0A151J434 A0A195FUE5 K7IMZ7 A0A232ELJ6 A0A151WIL1 A0A151HYP6 E0VSR2 A0A1S3J8R9 A0A1S3K505 E2BW17 A0A2A3E6E3 A0A2C9JQQ3 V3Z8D9 W4XCH6 C3Y8Q6 K1P6F0 R7UI93 A0A0C9PZJ6 F6S428 A0A1L8EQB6 W5M0E5 A0A1L8EY17 A0A0L7R775 A0JMW2 A0A1B6GW16 A0A2B4RBP0 A0A0L8G7D7 H2S9A9 A0A210QL21 A0A3P9B6Z3 A0A3P8RJF3 A0A3B3RXV3 A0A3Q2WTL2 I3JCA3 N6TFE4 U4U796 A0A2T7PY40 A0A3M6TCZ2 H3B2F2 W5LCW1 A0A3B3WNP8 A0A2G8KC10 A0A3B4EUY4 A0A087Y2B7 H2YGR6 W5UCV2 A0A3B4DSU1 A0A146RJF3 A0A147AU06 A0A3Q3GPL0 M3ZQM0 A0A3B3TZJ0 A0A146RM00 A0A146QY71 A0A3B3RXT1 A0A3B5AZ10 A0A3Q4GCM0 A0A1S3S1P0 F6Q2F8 A0A3Q1GP08 A0A3Q0S811 A0A1W2WEK6 A0A0F8CDB1 A0A3B5REA6 A0A096M6B0 A0A3B4U7X3 A0A0F8CYL3 A0A3Q1JUX1 A0A1B6CAU1 A0A3P8SES1 A0A3Q2XHN2 A0A1A8I9T1 A0A1A8N110 A0A1A8UI62 A0A1A7ZL32 A0A315WDI0 A0A1A8ERE0
Pubmed
27251284
23674305
19121390
22118469
26354079
24845553
+ More
28004739 29403074 24508170 20798317 21719571 30249741 18362917 19820115 21347285 20075255 28648823 20566863 15562597 23254933 18563158 22992520 20431018 27762356 21551351 28812685 25186727 29240929 23537049 30382153 9215903 25329095 29023486 23127152 23542700 12481130 15114417 25835551 29703783
28004739 29403074 24508170 20798317 21719571 30249741 18362917 19820115 21347285 20075255 28648823 20566863 15562597 23254933 18563158 22992520 20431018 27762356 21551351 28812685 25186727 29240929 23537049 30382153 9215903 25329095 29023486 23127152 23542700 12481130 15114417 25835551 29703783
EMBL
KT182637
ANG83464.1
HM358275
ADN86379.1
NWSH01001365
PCG71534.1
+ More
FP102339 CBH09268.1 KC469893 AGC92707.1 BABH01020220 BABH01020221 AGBW02008005 OWR54316.1 KQ459232 KPJ02619.1 ODYU01013053 SOQ59706.1 NEVH01003493 PNF40731.1 KK852980 KDR12976.1 GEZM01060241 GEZM01060234 JAV71170.1 PYGN01001777 PSN32857.1 KK107161 EZA56571.1 GL438128 EFN69531.1 GL888120 EGI66984.1 GEZM01060243 GEZM01060240 JAV71167.1 QOIP01000003 RLU25121.1 KQ971363 EFA07827.2 ADTU01024933 KQ977329 KYN03559.1 KQ980211 KYN17205.1 KQ981276 KYN43504.1 NNAY01003559 OXU19201.1 KQ983089 KYQ47575.1 KQ976719 KYM76745.1 DS235755 EEB16418.1 GL451091 EFN80049.1 KZ288353 PBC27333.1 KB202953 ESO87153.1 AAGJ04111103 GG666492 EEN62962.1 JH815900 EKC19157.1 AMQN01007697 KB301197 ELU05823.1 GBYB01006978 JAG76745.1 AAMC01119012 CM004483 OCT61558.1 AHAT01032348 CM004482 OCT64179.1 KQ414642 KOC66732.1 BC126027 AAI26028.1 GECZ01003140 JAS66629.1 LSMT01000818 PFX14219.1 KQ423493 KOF72788.1 NEDP02003110 OWF49420.1 AERX01023595 APGK01039948 KB740975 ENN76488.1 KB632186 ERL89774.1 PZQS01000001 PVD38342.1 RCHS01003840 RMX39272.1 AFYH01079148 AFYH01079149 AFYH01079150 AFYH01079151 AFYH01079152 AFYH01079153 AFYH01079154 AFYH01079155 AFYH01079156 AFYH01079157 MRZV01000710 PIK45499.1 AYCK01006546 JT412399 AHH39831.1 GCES01118155 JAQ68167.1 GCES01004265 JAR82058.1 GCES01116973 JAQ69349.1 GCES01125249 JAQ61073.1 EAAA01001812 KQ042264 KKF16648.1 KQ041123 KKF30508.1 GEDC01026903 JAS10395.1 HAED01007402 SBQ93614.1 HAEF01021418 SBR62577.1 HAEJ01007308 SBS47765.1 HADY01004673 SBP43158.1 NHOQ01000004 PWA33847.1 HAEB01002519 SBQ49046.1
FP102339 CBH09268.1 KC469893 AGC92707.1 BABH01020220 BABH01020221 AGBW02008005 OWR54316.1 KQ459232 KPJ02619.1 ODYU01013053 SOQ59706.1 NEVH01003493 PNF40731.1 KK852980 KDR12976.1 GEZM01060241 GEZM01060234 JAV71170.1 PYGN01001777 PSN32857.1 KK107161 EZA56571.1 GL438128 EFN69531.1 GL888120 EGI66984.1 GEZM01060243 GEZM01060240 JAV71167.1 QOIP01000003 RLU25121.1 KQ971363 EFA07827.2 ADTU01024933 KQ977329 KYN03559.1 KQ980211 KYN17205.1 KQ981276 KYN43504.1 NNAY01003559 OXU19201.1 KQ983089 KYQ47575.1 KQ976719 KYM76745.1 DS235755 EEB16418.1 GL451091 EFN80049.1 KZ288353 PBC27333.1 KB202953 ESO87153.1 AAGJ04111103 GG666492 EEN62962.1 JH815900 EKC19157.1 AMQN01007697 KB301197 ELU05823.1 GBYB01006978 JAG76745.1 AAMC01119012 CM004483 OCT61558.1 AHAT01032348 CM004482 OCT64179.1 KQ414642 KOC66732.1 BC126027 AAI26028.1 GECZ01003140 JAS66629.1 LSMT01000818 PFX14219.1 KQ423493 KOF72788.1 NEDP02003110 OWF49420.1 AERX01023595 APGK01039948 KB740975 ENN76488.1 KB632186 ERL89774.1 PZQS01000001 PVD38342.1 RCHS01003840 RMX39272.1 AFYH01079148 AFYH01079149 AFYH01079150 AFYH01079151 AFYH01079152 AFYH01079153 AFYH01079154 AFYH01079155 AFYH01079156 AFYH01079157 MRZV01000710 PIK45499.1 AYCK01006546 JT412399 AHH39831.1 GCES01118155 JAQ68167.1 GCES01004265 JAR82058.1 GCES01116973 JAQ69349.1 GCES01125249 JAQ61073.1 EAAA01001812 KQ042264 KKF16648.1 KQ041123 KKF30508.1 GEDC01026903 JAS10395.1 HAED01007402 SBQ93614.1 HAEF01021418 SBR62577.1 HAEJ01007308 SBS47765.1 HADY01004673 SBP43158.1 NHOQ01000004 PWA33847.1 HAEB01002519 SBQ49046.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000053268
UP000235965
UP000027135
+ More
UP000245037 UP000053097 UP000000311 UP000007755 UP000279307 UP000007266 UP000005205 UP000078542 UP000078492 UP000078541 UP000002358 UP000215335 UP000075809 UP000078540 UP000009046 UP000085678 UP000008237 UP000242457 UP000076420 UP000030746 UP000007110 UP000001554 UP000005408 UP000014760 UP000008143 UP000186698 UP000018468 UP000053825 UP000225706 UP000053454 UP000005226 UP000242188 UP000265160 UP000265100 UP000261540 UP000264840 UP000005207 UP000019118 UP000030742 UP000245119 UP000275408 UP000008672 UP000018467 UP000261480 UP000230750 UP000261460 UP000028760 UP000007875 UP000221080 UP000261440 UP000261660 UP000002852 UP000261500 UP000261400 UP000261580 UP000087266 UP000008144 UP000257200 UP000261340 UP000261420 UP000265040 UP000265080 UP000264820
UP000245037 UP000053097 UP000000311 UP000007755 UP000279307 UP000007266 UP000005205 UP000078542 UP000078492 UP000078541 UP000002358 UP000215335 UP000075809 UP000078540 UP000009046 UP000085678 UP000008237 UP000242457 UP000076420 UP000030746 UP000007110 UP000001554 UP000005408 UP000014760 UP000008143 UP000186698 UP000018468 UP000053825 UP000225706 UP000053454 UP000005226 UP000242188 UP000265160 UP000265100 UP000261540 UP000264840 UP000005207 UP000019118 UP000030742 UP000245119 UP000275408 UP000008672 UP000018467 UP000261480 UP000230750 UP000261460 UP000028760 UP000007875 UP000221080 UP000261440 UP000261660 UP000002852 UP000261500 UP000261400 UP000261580 UP000087266 UP000008144 UP000257200 UP000261340 UP000261420 UP000265040 UP000265080 UP000264820
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A173FDM6
E2GIU6
A0A2A4JIY0
D0AB73
L7WWA6
H9J807
+ More
A0A212FKQ7 A0A194QB14 A0A2H1X389 A0A2J7RIS0 A0A067R4R1 A0A1Y1LFQ5 A0A2P8XLI2 A0A026WLP7 E2AAI4 F4WFK7 A0A1Y1LJH1 A0A3L8DXE8 D6WZ28 A0A158NSI2 A0A195CTV0 A0A151J434 A0A195FUE5 K7IMZ7 A0A232ELJ6 A0A151WIL1 A0A151HYP6 E0VSR2 A0A1S3J8R9 A0A1S3K505 E2BW17 A0A2A3E6E3 A0A2C9JQQ3 V3Z8D9 W4XCH6 C3Y8Q6 K1P6F0 R7UI93 A0A0C9PZJ6 F6S428 A0A1L8EQB6 W5M0E5 A0A1L8EY17 A0A0L7R775 A0JMW2 A0A1B6GW16 A0A2B4RBP0 A0A0L8G7D7 H2S9A9 A0A210QL21 A0A3P9B6Z3 A0A3P8RJF3 A0A3B3RXV3 A0A3Q2WTL2 I3JCA3 N6TFE4 U4U796 A0A2T7PY40 A0A3M6TCZ2 H3B2F2 W5LCW1 A0A3B3WNP8 A0A2G8KC10 A0A3B4EUY4 A0A087Y2B7 H2YGR6 W5UCV2 A0A3B4DSU1 A0A146RJF3 A0A147AU06 A0A3Q3GPL0 M3ZQM0 A0A3B3TZJ0 A0A146RM00 A0A146QY71 A0A3B3RXT1 A0A3B5AZ10 A0A3Q4GCM0 A0A1S3S1P0 F6Q2F8 A0A3Q1GP08 A0A3Q0S811 A0A1W2WEK6 A0A0F8CDB1 A0A3B5REA6 A0A096M6B0 A0A3B4U7X3 A0A0F8CYL3 A0A3Q1JUX1 A0A1B6CAU1 A0A3P8SES1 A0A3Q2XHN2 A0A1A8I9T1 A0A1A8N110 A0A1A8UI62 A0A1A7ZL32 A0A315WDI0 A0A1A8ERE0
A0A212FKQ7 A0A194QB14 A0A2H1X389 A0A2J7RIS0 A0A067R4R1 A0A1Y1LFQ5 A0A2P8XLI2 A0A026WLP7 E2AAI4 F4WFK7 A0A1Y1LJH1 A0A3L8DXE8 D6WZ28 A0A158NSI2 A0A195CTV0 A0A151J434 A0A195FUE5 K7IMZ7 A0A232ELJ6 A0A151WIL1 A0A151HYP6 E0VSR2 A0A1S3J8R9 A0A1S3K505 E2BW17 A0A2A3E6E3 A0A2C9JQQ3 V3Z8D9 W4XCH6 C3Y8Q6 K1P6F0 R7UI93 A0A0C9PZJ6 F6S428 A0A1L8EQB6 W5M0E5 A0A1L8EY17 A0A0L7R775 A0JMW2 A0A1B6GW16 A0A2B4RBP0 A0A0L8G7D7 H2S9A9 A0A210QL21 A0A3P9B6Z3 A0A3P8RJF3 A0A3B3RXV3 A0A3Q2WTL2 I3JCA3 N6TFE4 U4U796 A0A2T7PY40 A0A3M6TCZ2 H3B2F2 W5LCW1 A0A3B3WNP8 A0A2G8KC10 A0A3B4EUY4 A0A087Y2B7 H2YGR6 W5UCV2 A0A3B4DSU1 A0A146RJF3 A0A147AU06 A0A3Q3GPL0 M3ZQM0 A0A3B3TZJ0 A0A146RM00 A0A146QY71 A0A3B3RXT1 A0A3B5AZ10 A0A3Q4GCM0 A0A1S3S1P0 F6Q2F8 A0A3Q1GP08 A0A3Q0S811 A0A1W2WEK6 A0A0F8CDB1 A0A3B5REA6 A0A096M6B0 A0A3B4U7X3 A0A0F8CYL3 A0A3Q1JUX1 A0A1B6CAU1 A0A3P8SES1 A0A3Q2XHN2 A0A1A8I9T1 A0A1A8N110 A0A1A8UI62 A0A1A7ZL32 A0A315WDI0 A0A1A8ERE0
Ontologies
KEGG
GO
PANTHER
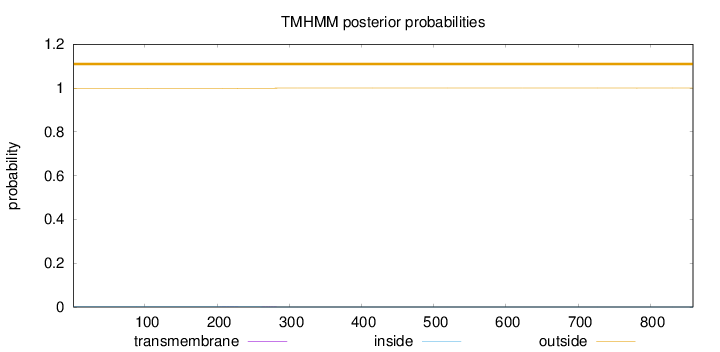
Topology
Length:
859
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0346700000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00152
outside
1 - 859
Population Genetic Test Statistics
Pi
178.626894
Theta
161.130997
Tajima's D
0.526802
CLR
0.14574
CSRT
0.523873806309684
Interpretation
Uncertain
