Pre Gene Modal
BGIBMGA005650
Annotation
poly(A)-specific_ribonuclease_[Bombyx_mori]
Full name
Poly(A)-specific ribonuclease PARN
Alternative Name
Deadenylating nuclease
Deadenylation nuclease
Polyadenylate-specific ribonuclease
parn-A
Deadenylation nuclease
Polyadenylate-specific ribonuclease
parn-A
Location in the cell
Nuclear Reliability : 2.042
Sequence
CDS
ATGGAAGTAATACGAAAAAATTTCAAAGAGGCATTGCCTCTTGCAGCAGCTTCTGTTAAGAAAGCGGACTTTCTAGTGATAGATACTGAGTTCACTGGCATTATAAACGGCCGGGATGTTTCGATGTTTGACACACCAGAGGAGTATTACAAATGCACTCAGAAAGGTTCTTCGGAATTTCTACTAATACAGTTTGGCCTGTGTGCTTTTCACTGGAATCATAAGGAGAAGCATTACATGAACGACGCATATAATTTTTACCTGTTTCCTCGTGGCAGCCCCGGCCCAGACAGAATGTTCATGTGTCAGAGTTCCAGCTTAGATTTCTTAGCTTCGCAGGGTTTTGATTTCAATAAATTAATAAAAGATGGAATCTCCTACATGACAGTACCTATAGAGTGCAAGCTAAGAGAAAACCTAACAGAGAGACAAAAGACATATTCAAAAGGAAAAGATTCCATTAAGGTACCAGATGAACATAAGGTGTATGTTGAAGACATTTGTAACCAAGTACACCAGTTTTTAGAAGAAGGACAGTTAGACGAAATGGAAATTGACAAGTGTAATTCATTTGTTAGATTATTACTGTTTCAAGAGTTGAGAGCTCGTTTTAAAGACAAAATCTTGGTCGAAACTAAGATTCTAGAAAATAAAAACAGATTATTAAATGTAAAAAGAGTCAAGTCACAGGATGATCTCAAGAACCATGATGCTCTAAAGAAGGAACGAGAGTGGGAAGATTTTGAAGATGCTGTCGGCTTCTCAAAAGTAGCTAGGATGATAAGCGAGTCTGAGAAACTGGTGGTCGGACACAACATGTTACTGGACGTAATGCACACATTGAATCATTTCTTCCAGCCTTTACCTGCAGAATACACCCAGTTCAAAGAGTTTACACACTGCATGTTCCCACATATCTTAGACACAAAATACATGTCAAGTTTGCCTCCATTTAAAGATAAGGTGAATTCAAGCATTCTGAAACATTTGTTGGCAACACTGTCAGCTGCGCCATTTTCTCTGCCTAAAGTTGTGTCCGATGAAGGTCGAGGTTACAGTCAACTCCACGAGAAGCACCACGAAGCCGGCTACGACGCCTACGTCACGGGGCTCTGCTTCCTGGCCATGCACTCGCACCTCGCCAACATGAGGGGCGACGACACCACAAGGTTCCTGTCCATCACGTCGCCGCTCATCAAGCCCTTCCTGAACAAGGTGTTCCTGTCCAGGACGGCTCATCAGGACTCGCCGTTCATAAACCTGGCGGGACCGGAACCACTGCCTCCCAGAGACCACGTGTTTCATTTGGCGTTTCCCAGAGAGTGGCAAAGGAATGAAATCACGCAACTATTTAGTCCGTTTGGTCCAATAACAGTACAATTCATCGATGATACGTCAGCGTTCGTGGCGTTAGCGCGACGTGAACAGGCGACGTCTGTTAGCAAAGCACTCGCGAAGCATTCCAAGATAACCCTCACATCGTATTACAAGTATAAGAAGATTAATGATACGCCCAGCAGTAAGATAACGACGAAAACGGCGGAGGCTGCGAACAACGACTGCAAGAAAACGTATGCAATACTCATTATATCATTAGTGGTAATTTTAATTGCATTCTTCCTGCGTGGCTTCAATAAGCTATACTGA
Protein
MEVIRKNFKEALPLAAASVKKADFLVIDTEFTGIINGRDVSMFDTPEEYYKCTQKGSSEFLLIQFGLCAFHWNHKEKHYMNDAYNFYLFPRGSPGPDRMFMCQSSSLDFLASQGFDFNKLIKDGISYMTVPIECKLRENLTERQKTYSKGKDSIKVPDEHKVYVEDICNQVHQFLEEGQLDEMEIDKCNSFVRLLLFQELRARFKDKILVETKILENKNRLLNVKRVKSQDDLKNHDALKKEREWEDFEDAVGFSKVARMISESEKLVVGHNMLLDVMHTLNHFFQPLPAEYTQFKEFTHCMFPHILDTKYMSSLPPFKDKVNSSILKHLLATLSAAPFSLPKVVSDEGRGYSQLHEKHHEAGYDAYVTGLCFLAMHSHLANMRGDDTTRFLSITSPLIKPFLNKVFLSRTAHQDSPFINLAGPEPLPPRDHVFHLAFPREWQRNEITQLFSPFGPITVQFIDDTSAFVALARREQATSVSKALAKHSKITLTSYYKYKKINDTPSSKITTKTAEAANNDCKKTYAILIISLVVILIAFFLRGFNKLY
Summary
Description
3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs. Required during meiotic maturation to silence certain maternal mRNAs translationally. Does not require an adenosine residue at the 3' end, however, the addition of 25 non-adenylate residues at the 3' terminus, or a 3' terminal phosphate is inhibitory. Involved in dormant mRNAs regulation during oocyte maturation by counteracting polyadenylation mediated by papd4/gld2nt in immature eggs. During maturation it is excluded from the ribonucleoprotein complex, allowing poly(A) elongation by papd4/gld2nt and activation of mRNAs.
3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development. Involved in nonsense-mediated mRNA decay, a critical process of selective degradation of mRNAs that contain premature stop codons. Also involved in degradation of inherently unstable mRNAs that contain AU-rich elements (AREs) in their 3'-UTR, possibly via its interaction with KHSRP. Probably mediates the removal of poly(A) tails of AREs mRNAs, which constitutes the first step of destabilization (By similarity). Interacts with both the 3'-end poly(A) tail and the 5'-end cap structure during degradation, the interaction with the cap structure being required for an efficient degradation of poly(A) tails (By similarity) (PubMed:10698948, PubMed:9736620). Also able to recognize poly(A) tails of microRNAs such as MIR21 and H/ACA box snoRNAs (small nucleolar RNAs) leading to microRNAs degradation or snoRNA increased stability (By similarity).
3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development. Interacts with both the 3'-end poly(A) tail and the 5'-end cap structure during degradation, the interaction with the cap structure being required for an efficient degradation of poly(A) tails. Involved in nonsense-mediated mRNA decay, a critical process of selective degradation of mRNAs that contain premature stop codons. Also involved in degradation of inherently unstable mRNAs that contain AU-rich elements (AREs) in their 3'-UTR, possibly via its interaction with KHSRP. Probably mediates the removal of poly(A) tails of AREs mRNAs, which constitutes the first step of destabilization (PubMed:10882133, PubMed:11359775, PubMed:12748283, PubMed:15175153, PubMed:9736620). Also able to recognize and trim poly(A) tails of microRNAs such as MIR21 and H/ACA box snoRNAs (small nucleolar RNAs) leading to microRNAs degradation or snoRNA increased stability (PubMed:25049417, PubMed:22442037).
3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development. Interacts with both the 3'-end poly(A) tail and the 5'-end cap structure during degradation, the interaction with the cap structure being required for an efficient degradation of poly(A) tails. Involved in nonsense-mediated mRNA decay, a critical process of selective degradation of mRNAs that contain premature stop codons. Also involved in degradation of inherently unstable mRNAs that contain AU-rich elements (AREs) in their 3'-UTR, possibly via its interaction with KHSRP. Probably mediates the removal of poly(A) tails of AREs mRNAs, which constitutes the first step of destabilization (By similarity). Also able to recognize poly(A) tails of microRNAs such as MIR21 and H/ACA box snoRNAs (small nucleolar RNAs) leading to microRNAs degradation or snoRNA increased stability (By similarity).
3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development. Involved in nonsense-mediated mRNA decay, a critical process of selective degradation of mRNAs that contain premature stop codons. Also involved in degradation of inherently unstable mRNAs that contain AU-rich elements (AREs) in their 3'-UTR, possibly via its interaction with KHSRP. Probably mediates the removal of poly(A) tails of AREs mRNAs, which constitutes the first step of destabilization (By similarity). Interacts with both the 3'-end poly(A) tail and the 5'-end cap structure during degradation, the interaction with the cap structure being required for an efficient degradation of poly(A) tails (By similarity) (PubMed:10698948, PubMed:9736620). Also able to recognize poly(A) tails of microRNAs such as MIR21 and H/ACA box snoRNAs (small nucleolar RNAs) leading to microRNAs degradation or snoRNA increased stability (By similarity).
3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development. Interacts with both the 3'-end poly(A) tail and the 5'-end cap structure during degradation, the interaction with the cap structure being required for an efficient degradation of poly(A) tails. Involved in nonsense-mediated mRNA decay, a critical process of selective degradation of mRNAs that contain premature stop codons. Also involved in degradation of inherently unstable mRNAs that contain AU-rich elements (AREs) in their 3'-UTR, possibly via its interaction with KHSRP. Probably mediates the removal of poly(A) tails of AREs mRNAs, which constitutes the first step of destabilization (PubMed:10882133, PubMed:11359775, PubMed:12748283, PubMed:15175153, PubMed:9736620). Also able to recognize and trim poly(A) tails of microRNAs such as MIR21 and H/ACA box snoRNAs (small nucleolar RNAs) leading to microRNAs degradation or snoRNA increased stability (PubMed:25049417, PubMed:22442037).
3'-exoribonuclease that has a preference for poly(A) tails of mRNAs, thereby efficiently degrading poly(A) tails. Exonucleolytic degradation of the poly(A) tail is often the first step in the decay of eukaryotic mRNAs and is also used to silence certain maternal mRNAs translationally during oocyte maturation and early embryonic development. Interacts with both the 3'-end poly(A) tail and the 5'-end cap structure during degradation, the interaction with the cap structure being required for an efficient degradation of poly(A) tails. Involved in nonsense-mediated mRNA decay, a critical process of selective degradation of mRNAs that contain premature stop codons. Also involved in degradation of inherently unstable mRNAs that contain AU-rich elements (AREs) in their 3'-UTR, possibly via its interaction with KHSRP. Probably mediates the removal of poly(A) tails of AREs mRNAs, which constitutes the first step of destabilization (By similarity). Also able to recognize poly(A) tails of microRNAs such as MIR21 and H/ACA box snoRNAs (small nucleolar RNAs) leading to microRNAs degradation or snoRNA increased stability (By similarity).
Catalytic Activity
Exonucleolytic cleavage of poly(A) to 5'-AMP.
Cofactor
a divalent metal cation
Mg(2+)
Mg(2+)
Subunit
Component of a complex at least composed of cpeb1, cpsf1, papd4/gld2, pabpc1/ePAB, parn and sympk.
Homodimer. Found in a mRNA decay complex with RENT1, RENT2 and RENT3B. Interacts with KHSRP. Interacts with CELF1/CUGBP1. Interacts with ZC3HAV1 in an RNA-independent manner. Interacts with DHX36.
Homodimer (PubMed:10801819, PubMed:16281054). Found in a mRNA decay complex with RENT1, RENT2 and RENT3B (PubMed:14527413). Interacts with KHSRP (PubMed:15175153). Interacts with CELF1/CUGBP1 (PubMed:16601207). Interacts with ZC3HAV1 in an RNA-independent manner (PubMed:21876179). Interacts with DHX36 (PubMed:14731398).
Homodimer. Found in a mRNA decay complex with RENT1, RENT2 and RENT3B. Interacts with KHSRP. Interacts with CELF1/CUGBP1. Interacts with ZC3HAV1 in an RNA-independent manner. Interacts with DHX36.
Homodimer (PubMed:10801819, PubMed:16281054). Found in a mRNA decay complex with RENT1, RENT2 and RENT3B (PubMed:14527413). Interacts with KHSRP (PubMed:15175153). Interacts with CELF1/CUGBP1 (PubMed:16601207). Interacts with ZC3HAV1 in an RNA-independent manner (PubMed:21876179). Interacts with DHX36 (PubMed:14731398).
Similarity
Belongs to the CAF1 family.
Keywords
Cytoplasm
Direct protein sequencing
Exonuclease
Hydrolase
Meiosis
Metal-binding
Nuclease
Nucleus
RNA-binding
Acetylation
Complete proteome
Magnesium
Nonsense-mediated mRNA decay
Phosphoprotein
Reference proteome
3D-structure
Alternative splicing
Disease mutation
Dyskeratosis congenita
Feature
chain Poly(A)-specific ribonuclease PARN
splice variant In isoform 2.
sequence variant In DKCB6; dbSNP:rs786200999.
splice variant In isoform 2.
sequence variant In DKCB6; dbSNP:rs786200999.
Uniprot
H9J808
B9X256
A0A2A4JHI6
A0A3S2NI25
A0A173FD15
D0AB72
+ More
A0A212FKP2 L7X1H3 A0A2H1VJ40 A0A194QAQ6 A0A1E1W4W7 A0A2J7QAK1 A0A067R0F3 A0A1S3JBU0 T1JBS2 A0A1S3K3F2 A0A2P8YJZ6 E9IBK7 A0A087ZTK8 A0A1Y1LHM7 A0A026WYM1 A0A226MLI7 A0A3L8D835 A0A2A3E552 B5DFR2 A0A0M9A5A4 Q07G83 D6WYW0 F6UAK5 A0A1L8EQ87 A0A1L8EXJ1 Q90ZA1 K7IX58 A0A2P4TFY5 E2AZW9 A0A232F832 W5PCG5 A0A2U9BNZ1 A0A3B4ZT46 U3D8R3 F1NEI8 E2C8P3 A0A093G7B5 G3TEC9 P69341 A0A2J8R143 A0A0D9R712 A0A2K6EAH7 S4RGQ4 A0A2Y9DES8 A0A2J8JL49 I3IUA4 A0A099Z7R2 H9FV31 W5U5L5 G3RWB8 A0A2Y9QXV2 O95453 G1LXQ5 A0A2R9APG9 K7BWE4 A0A250Y414 Q5RC51 A0A0S3NU58 A0A3B4UNQ9 W5LXH8 G1RH04 F6Q3J7 I3M9X4 A0A384CJZ2 A0A3Q7XN02 A0A2K5E2Q6 A0A3Q2LEK8 A0A2K5RAN0 I3IUA3 A0A337S613 A0A2T7PXW8 W5LXE4 F6XLZ1 H3CS48 A0A0L7R0U2 A0A2U4ATD1 A0A287AI76 M3YSP4 A0A1S3WJL1 K7G9L5 A0A1S3ES48 G3Q7C0 A0A341BBS8 A0A2Y9LRG7 A0A2Y9KH53 F6XU41 A0A341BF47 A0A3Q0D5Z0 A0A3Q3LTX5 A0A340Y131 A0A3P9C7U1 A0A3Q3A9J3
A0A212FKP2 L7X1H3 A0A2H1VJ40 A0A194QAQ6 A0A1E1W4W7 A0A2J7QAK1 A0A067R0F3 A0A1S3JBU0 T1JBS2 A0A1S3K3F2 A0A2P8YJZ6 E9IBK7 A0A087ZTK8 A0A1Y1LHM7 A0A026WYM1 A0A226MLI7 A0A3L8D835 A0A2A3E552 B5DFR2 A0A0M9A5A4 Q07G83 D6WYW0 F6UAK5 A0A1L8EQ87 A0A1L8EXJ1 Q90ZA1 K7IX58 A0A2P4TFY5 E2AZW9 A0A232F832 W5PCG5 A0A2U9BNZ1 A0A3B4ZT46 U3D8R3 F1NEI8 E2C8P3 A0A093G7B5 G3TEC9 P69341 A0A2J8R143 A0A0D9R712 A0A2K6EAH7 S4RGQ4 A0A2Y9DES8 A0A2J8JL49 I3IUA4 A0A099Z7R2 H9FV31 W5U5L5 G3RWB8 A0A2Y9QXV2 O95453 G1LXQ5 A0A2R9APG9 K7BWE4 A0A250Y414 Q5RC51 A0A0S3NU58 A0A3B4UNQ9 W5LXH8 G1RH04 F6Q3J7 I3M9X4 A0A384CJZ2 A0A3Q7XN02 A0A2K5E2Q6 A0A3Q2LEK8 A0A2K5RAN0 I3IUA3 A0A337S613 A0A2T7PXW8 W5LXE4 F6XLZ1 H3CS48 A0A0L7R0U2 A0A2U4ATD1 A0A287AI76 M3YSP4 A0A1S3WJL1 K7G9L5 A0A1S3ES48 G3Q7C0 A0A341BBS8 A0A2Y9LRG7 A0A2Y9KH53 F6XU41 A0A341BF47 A0A3Q0D5Z0 A0A3Q3LTX5 A0A340Y131 A0A3P9C7U1 A0A3Q3A9J3
EC Number
3.1.13.4
Pubmed
19121390
27251284
22118469
23674305
26354079
24845553
+ More
29403074 21282665 28004739 24508170 30249741 18362917 19820115 20431018 27762356 11424938 9736620 12573214 17052452 20075255 20798317 28648823 20809919 25243066 15592404 9099687 10698948 25186727 17431167 25319552 23127152 22398555 14702039 15616553 15489334 10640832 10801819 10882133 11359775 11742007 12429849 14527413 12748283 14731398 15175153 15358788 16964243 16601207 18669648 19413330 19690332 19608861 20932473 20068231 21269460 21876179 21406692 22442037 23186163 24275569 25049417 25893599 25848748 16281054 20010809 22722832 16136131 28087693 26514418 19892987 24813606 17975172 15496914 17381049 16341006 28071753
29403074 21282665 28004739 24508170 30249741 18362917 19820115 20431018 27762356 11424938 9736620 12573214 17052452 20075255 20798317 28648823 20809919 25243066 15592404 9099687 10698948 25186727 17431167 25319552 23127152 22398555 14702039 15616553 15489334 10640832 10801819 10882133 11359775 11742007 12429849 14527413 12748283 14731398 15175153 15358788 16964243 16601207 18669648 19413330 19690332 19608861 20932473 20068231 21269460 21876179 21406692 22442037 23186163 24275569 25049417 25893599 25848748 16281054 20010809 22722832 16136131 28087693 26514418 19892987 24813606 17975172 15496914 17381049 16341006 28071753
EMBL
BABH01020223
AB485782
BAH23570.1
NWSH01001365
PCG71535.1
RSAL01000090
+ More
RVE48111.1 KT182637 ANG83463.1 FP102339 CBH09267.1 AGBW02008005 OWR54317.1 KC469893 AGC92706.2 ODYU01002851 SOQ40847.1 KQ459232 KPJ02618.1 GDQN01009050 JAT82004.1 NEVH01016330 PNF25589.1 KK852804 KDR16211.1 JH432018 PYGN01000541 PSN44566.1 GL762111 EFZ22121.1 GEZM01055097 JAV73149.1 KK107078 EZA60239.1 MCFN01000669 OXB56142.1 QOIP01000011 RLU16444.1 KZ288368 PBC26818.1 BC169159 AAI69159.1 KQ435732 KOX77527.1 CR760641 CAL49352.1 KQ971363 EFA07855.1 AAMC01046567 AAMC01046568 AAMC01046569 AAMC01046570 AAMC01046571 AAMC01046572 AAMC01046573 CM004483 OCT61470.1 CM004482 OCT64052.1 AF309688 BC068919 BC073682 AAZX01000623 PPHD01000639 POI35266.1 GL444280 EFN61018.1 NNAY01000683 OXU26994.1 AMGL01068514 AMGL01068515 AMGL01068516 AMGL01068517 AMGL01068518 AMGL01068519 CP026250 AWP05326.1 GAMT01002170 GAMS01007066 GAMR01005114 GAMQ01002084 GAMP01008800 JAB09691.1 JAB16070.1 JAB28818.1 JAB39767.1 JAB43955.1 AADN05000376 GL453666 EFN75773.1 KL215669 KFV66105.1 BC150015 NDHI03003836 PNJ02239.1 AQIB01118291 AQIB01118292 AQIB01118293 AQIB01118294 AQIB01118295 AQIB01118296 AQIB01118297 AQIB01118298 AQIB01118299 NBAG03000447 PNI23493.1 AERX01047414 KL889709 KGL77093.1 JSUE03025737 JSUE03025738 JSUE03025739 JSUE03025740 JU334737 JU472194 JV045146 AFE78490.1 AFH28998.1 AFI35217.1 JT405467 AHH37193.1 CABD030099076 CABD030099077 CABD030099078 CABD030099079 CABD030099080 CABD030099081 CABD030099082 CABD030099083 CABD030099084 CABD030099085 AJ005698 AK293189 AK299653 AK301648 AK315020 AC009167 AC092291 KF456163 CH471112 BC050029 ACTA01011153 ACTA01019153 ACTA01027153 ACTA01035153 ACTA01043152 AJFE02117450 AJFE02117451 AJFE02117452 AJFE02117453 AJFE02117454 AJFE02117455 AJFE02117456 AJFE02117457 AJFE02117458 AJFE02117459 AACZ04058832 AC192135 GABC01005196 GABF01005845 GABD01010747 GABE01002844 GABE01002843 JAA06142.1 JAA16300.1 JAA22353.1 JAA41895.1 GFFV01001666 JAV38279.1 CR858430 AB984736 LC192268 BAT46600.1 BBG62308.1 AHAT01032371 ADFV01049477 ADFV01049478 ADFV01049479 ADFV01049480 ADFV01049481 ADFV01049482 ADFV01049483 ADFV01049484 ADFV01049485 ADFV01049486 AGTP01030790 AGTP01030791 AGTP01030792 AGTP01030793 AGTP01030794 AANG04001668 PZQS01000001 PVD38240.1 KQ414669 KOC64462.1 AEMK02000018 AEYP01027405 AEYP01027406 AEYP01027407 AEYP01027408 AEYP01027409 AEYP01027410 AEYP01027411 AEYP01027412 AEYP01027413 AEYP01027414 AGCU01158713 AGCU01158714 AGCU01158715 AGCU01158716 AGCU01158717 AGCU01158718 AGCU01158719 AGCU01158720 AGCU01158721 AGCU01158722 AAEX03004504 AAEX03004505
RVE48111.1 KT182637 ANG83463.1 FP102339 CBH09267.1 AGBW02008005 OWR54317.1 KC469893 AGC92706.2 ODYU01002851 SOQ40847.1 KQ459232 KPJ02618.1 GDQN01009050 JAT82004.1 NEVH01016330 PNF25589.1 KK852804 KDR16211.1 JH432018 PYGN01000541 PSN44566.1 GL762111 EFZ22121.1 GEZM01055097 JAV73149.1 KK107078 EZA60239.1 MCFN01000669 OXB56142.1 QOIP01000011 RLU16444.1 KZ288368 PBC26818.1 BC169159 AAI69159.1 KQ435732 KOX77527.1 CR760641 CAL49352.1 KQ971363 EFA07855.1 AAMC01046567 AAMC01046568 AAMC01046569 AAMC01046570 AAMC01046571 AAMC01046572 AAMC01046573 CM004483 OCT61470.1 CM004482 OCT64052.1 AF309688 BC068919 BC073682 AAZX01000623 PPHD01000639 POI35266.1 GL444280 EFN61018.1 NNAY01000683 OXU26994.1 AMGL01068514 AMGL01068515 AMGL01068516 AMGL01068517 AMGL01068518 AMGL01068519 CP026250 AWP05326.1 GAMT01002170 GAMS01007066 GAMR01005114 GAMQ01002084 GAMP01008800 JAB09691.1 JAB16070.1 JAB28818.1 JAB39767.1 JAB43955.1 AADN05000376 GL453666 EFN75773.1 KL215669 KFV66105.1 BC150015 NDHI03003836 PNJ02239.1 AQIB01118291 AQIB01118292 AQIB01118293 AQIB01118294 AQIB01118295 AQIB01118296 AQIB01118297 AQIB01118298 AQIB01118299 NBAG03000447 PNI23493.1 AERX01047414 KL889709 KGL77093.1 JSUE03025737 JSUE03025738 JSUE03025739 JSUE03025740 JU334737 JU472194 JV045146 AFE78490.1 AFH28998.1 AFI35217.1 JT405467 AHH37193.1 CABD030099076 CABD030099077 CABD030099078 CABD030099079 CABD030099080 CABD030099081 CABD030099082 CABD030099083 CABD030099084 CABD030099085 AJ005698 AK293189 AK299653 AK301648 AK315020 AC009167 AC092291 KF456163 CH471112 BC050029 ACTA01011153 ACTA01019153 ACTA01027153 ACTA01035153 ACTA01043152 AJFE02117450 AJFE02117451 AJFE02117452 AJFE02117453 AJFE02117454 AJFE02117455 AJFE02117456 AJFE02117457 AJFE02117458 AJFE02117459 AACZ04058832 AC192135 GABC01005196 GABF01005845 GABD01010747 GABE01002844 GABE01002843 JAA06142.1 JAA16300.1 JAA22353.1 JAA41895.1 GFFV01001666 JAV38279.1 CR858430 AB984736 LC192268 BAT46600.1 BBG62308.1 AHAT01032371 ADFV01049477 ADFV01049478 ADFV01049479 ADFV01049480 ADFV01049481 ADFV01049482 ADFV01049483 ADFV01049484 ADFV01049485 ADFV01049486 AGTP01030790 AGTP01030791 AGTP01030792 AGTP01030793 AGTP01030794 AANG04001668 PZQS01000001 PVD38240.1 KQ414669 KOC64462.1 AEMK02000018 AEYP01027405 AEYP01027406 AEYP01027407 AEYP01027408 AEYP01027409 AEYP01027410 AEYP01027411 AEYP01027412 AEYP01027413 AEYP01027414 AGCU01158713 AGCU01158714 AGCU01158715 AGCU01158716 AGCU01158717 AGCU01158718 AGCU01158719 AGCU01158720 AGCU01158721 AGCU01158722 AAEX03004504 AAEX03004505
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053268
UP000235965
+ More
UP000027135 UP000085678 UP000245037 UP000005203 UP000053097 UP000198323 UP000279307 UP000242457 UP000053105 UP000007266 UP000008143 UP000186698 UP000002358 UP000000311 UP000215335 UP000002356 UP000246464 UP000261400 UP000008225 UP000000539 UP000008237 UP000053875 UP000007646 UP000009136 UP000029965 UP000233120 UP000245300 UP000248480 UP000005207 UP000053641 UP000006718 UP000001519 UP000005640 UP000008912 UP000240080 UP000002277 UP000001595 UP000261420 UP000018468 UP000001073 UP000002281 UP000005215 UP000261680 UP000291021 UP000286642 UP000233020 UP000233040 UP000011712 UP000245119 UP000007303 UP000053825 UP000245320 UP000008227 UP000000715 UP000079721 UP000007267 UP000081671 UP000007635 UP000252040 UP000248483 UP000248482 UP000002254 UP000189706 UP000261660 UP000265300 UP000265160 UP000264800
UP000027135 UP000085678 UP000245037 UP000005203 UP000053097 UP000198323 UP000279307 UP000242457 UP000053105 UP000007266 UP000008143 UP000186698 UP000002358 UP000000311 UP000215335 UP000002356 UP000246464 UP000261400 UP000008225 UP000000539 UP000008237 UP000053875 UP000007646 UP000009136 UP000029965 UP000233120 UP000245300 UP000248480 UP000005207 UP000053641 UP000006718 UP000001519 UP000005640 UP000008912 UP000240080 UP000002277 UP000001595 UP000261420 UP000018468 UP000001073 UP000002281 UP000005215 UP000261680 UP000291021 UP000286642 UP000233020 UP000233040 UP000011712 UP000245119 UP000007303 UP000053825 UP000245320 UP000008227 UP000000715 UP000079721 UP000007267 UP000081671 UP000007635 UP000252040 UP000248483 UP000248482 UP000002254 UP000189706 UP000261660 UP000265300 UP000265160 UP000264800
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J808
B9X256
A0A2A4JHI6
A0A3S2NI25
A0A173FD15
D0AB72
+ More
A0A212FKP2 L7X1H3 A0A2H1VJ40 A0A194QAQ6 A0A1E1W4W7 A0A2J7QAK1 A0A067R0F3 A0A1S3JBU0 T1JBS2 A0A1S3K3F2 A0A2P8YJZ6 E9IBK7 A0A087ZTK8 A0A1Y1LHM7 A0A026WYM1 A0A226MLI7 A0A3L8D835 A0A2A3E552 B5DFR2 A0A0M9A5A4 Q07G83 D6WYW0 F6UAK5 A0A1L8EQ87 A0A1L8EXJ1 Q90ZA1 K7IX58 A0A2P4TFY5 E2AZW9 A0A232F832 W5PCG5 A0A2U9BNZ1 A0A3B4ZT46 U3D8R3 F1NEI8 E2C8P3 A0A093G7B5 G3TEC9 P69341 A0A2J8R143 A0A0D9R712 A0A2K6EAH7 S4RGQ4 A0A2Y9DES8 A0A2J8JL49 I3IUA4 A0A099Z7R2 H9FV31 W5U5L5 G3RWB8 A0A2Y9QXV2 O95453 G1LXQ5 A0A2R9APG9 K7BWE4 A0A250Y414 Q5RC51 A0A0S3NU58 A0A3B4UNQ9 W5LXH8 G1RH04 F6Q3J7 I3M9X4 A0A384CJZ2 A0A3Q7XN02 A0A2K5E2Q6 A0A3Q2LEK8 A0A2K5RAN0 I3IUA3 A0A337S613 A0A2T7PXW8 W5LXE4 F6XLZ1 H3CS48 A0A0L7R0U2 A0A2U4ATD1 A0A287AI76 M3YSP4 A0A1S3WJL1 K7G9L5 A0A1S3ES48 G3Q7C0 A0A341BBS8 A0A2Y9LRG7 A0A2Y9KH53 F6XU41 A0A341BF47 A0A3Q0D5Z0 A0A3Q3LTX5 A0A340Y131 A0A3P9C7U1 A0A3Q3A9J3
A0A212FKP2 L7X1H3 A0A2H1VJ40 A0A194QAQ6 A0A1E1W4W7 A0A2J7QAK1 A0A067R0F3 A0A1S3JBU0 T1JBS2 A0A1S3K3F2 A0A2P8YJZ6 E9IBK7 A0A087ZTK8 A0A1Y1LHM7 A0A026WYM1 A0A226MLI7 A0A3L8D835 A0A2A3E552 B5DFR2 A0A0M9A5A4 Q07G83 D6WYW0 F6UAK5 A0A1L8EQ87 A0A1L8EXJ1 Q90ZA1 K7IX58 A0A2P4TFY5 E2AZW9 A0A232F832 W5PCG5 A0A2U9BNZ1 A0A3B4ZT46 U3D8R3 F1NEI8 E2C8P3 A0A093G7B5 G3TEC9 P69341 A0A2J8R143 A0A0D9R712 A0A2K6EAH7 S4RGQ4 A0A2Y9DES8 A0A2J8JL49 I3IUA4 A0A099Z7R2 H9FV31 W5U5L5 G3RWB8 A0A2Y9QXV2 O95453 G1LXQ5 A0A2R9APG9 K7BWE4 A0A250Y414 Q5RC51 A0A0S3NU58 A0A3B4UNQ9 W5LXH8 G1RH04 F6Q3J7 I3M9X4 A0A384CJZ2 A0A3Q7XN02 A0A2K5E2Q6 A0A3Q2LEK8 A0A2K5RAN0 I3IUA3 A0A337S613 A0A2T7PXW8 W5LXE4 F6XLZ1 H3CS48 A0A0L7R0U2 A0A2U4ATD1 A0A287AI76 M3YSP4 A0A1S3WJL1 K7G9L5 A0A1S3ES48 G3Q7C0 A0A341BBS8 A0A2Y9LRG7 A0A2Y9KH53 F6XU41 A0A341BF47 A0A3Q0D5Z0 A0A3Q3LTX5 A0A340Y131 A0A3P9C7U1 A0A3Q3A9J3
PDB
3D45
E-value=1.11953e-109,
Score=1015
Ontologies
GO
GO:0003723
GO:0004535
GO:0006402
GO:0046872
GO:0005634
GO:0005737
GO:0016021
GO:0003676
GO:0000175
GO:0000289
GO:0043169
GO:0051321
GO:0019901
GO:0032212
GO:0051973
GO:0005730
GO:0010587
GO:0071051
GO:0110008
GO:0000495
GO:0090669
GO:1904872
GO:0000184
GO:0005829
GO:0070034
GO:0007292
GO:0043488
GO:0003730
GO:0004518
GO:0009451
GO:0090503
GO:0000166
GO:0003824
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleolus Some nuclear fraction is nucleolar. With evidence from 1 publications.
Nucleus
Nucleolus Some nuclear fraction is nucleolar. With evidence from 1 publications.
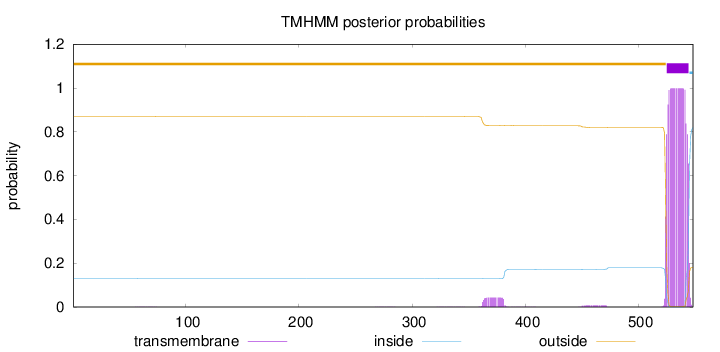
Length:
548
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.71512
Exp number, first 60 AAs:
0.005
Total prob of N-in:
0.13041
outside
1 - 524
TMhelix
525 - 544
inside
545 - 548
Population Genetic Test Statistics
Pi
225.321666
Theta
172.86612
Tajima's D
0.955761
CLR
0.149072
CSRT
0.648267586620669
Interpretation
Uncertain
