Gene
KWMTBOMO10119
Pre Gene Modal
BGIBMGA005652
Annotation
hypothetical_protein_KGM_00350_[Danaus_plexippus]
Full name
Protein cortex
Location in the cell
PlasmaMembrane Reliability : 2.466
Sequence
CDS
ATGCAAAACATATCTTGCTTGACTTCCTCAGTTCCAGAGCTTTTGGACTGGAGCAGTGATAACATTCTGGTGGCAGCTCTGGGCAGGAACTACCATAAATGGAGCTGGCGCACTCAAAGCCTTACCAACCAGGGTTTTACGCAATATGAAATACAATGCTGCAAGTTTGATCCTAGAGGAGAACTCTTACTGCTTGGCACTGACAACCAGATAACGGAGATTCACAACAACGCGATGAGCAAGTGGGTGGCTGAATGTTCCTGTTTGTGTAACAGATTTGTAGCCATCTGCAGCATCACTGCCGTCGATTGGAGCCCTACAGGAAACTCTTTTATAACGGGATGTTCTCGTGGAGTGCTGGTGTCATACACTCGCACTGGAGACATGCTGGGGTGGCGACGTGTGGGCGAGACGATGCTGGCGGTGCGCGTATCTCCGGACGCACGGTATGTGGCGGCCGCGGCCGTCAACGGAACCACCGTACTCGTCATGACCTGGCCGCTACTCCACTTGTACTCTAGTTTGGGCTCTAACTGGACTGTTAAGGCTTTGAGCTGGCACCCATGGCGCTCCGCCTTGCTGGGTGTGGGCGCAGTGACGTCACGGATGAACGCGCACGTCGCGGTGTGGGACGCGCCCACCTCCCGGGTCACGGACACCACAATCCGCCACAGCCGCCATAGTCTGGACGCTATGCTCTTCAGCCATAAGACCGGAGAGCTGGTCATCAGCTTGTGGAACCCGGAACGAGCCGTTTCATCGCCGAAGACTTGTTCACAATTGGTAGTGATGAGTGGACCAGATAAAGTGGTCGATCAATGGGGCGAAGAACGCTCGGGACTGGACCGGATCAGGACTATGGTTTTCAGTCCCGATGGAACTAAGCTTGCGACAGCAACAGCAGACGAAGACCTCATCATCTGGAACTTTCTCCCGGAAGACTCCAAGAGGAAGAAGTGGAAATGCGACAGATTCTGGGCGCTTCCGCTGTACCTGGACGAAACGACTTACGGCCTCTCAATGCGTTAG
Protein
MQNISCLTSSVPELLDWSSDNILVAALGRNYHKWSWRTQSLTNQGFTQYEIQCCKFDPRGELLLLGTDNQITEIHNNAMSKWVAECSCLCNRFVAICSITAVDWSPTGNSFITGCSRGVLVSYTRTGDMLGWRRVGETMLAVRVSPDARYVAAAAVNGTTVLVMTWPLLHLYSSLGSNWTVKALSWHPWRSALLGVGAVTSRMNAHVAVWDAPTSRVTDTTIRHSRHSLDAMLFSHKTGELVISLWNPERAVSSPKTCSQLVVMSGPDKVVDQWGEERSGLDRIRTMVFSPDGTKLATATADEDLIIWNFLPEDSKRKKWKCDRFWALPLYLDETTYGLSMR
Summary
Description
Controls wing pigmentation patterning by regulating scale cell development, thereby playing a key role in mimicry and crypsis (PubMed:27251285). Probably acts as an activator of the anaphase promoting complex/cyclosome (APC/C) that promotes the ubiquitin ligase activity and substrate specificity of the APC/C (By similarity).
Controls wing pigmentation patterning, possibly by regulating scale cell development (PubMed:27251284). Probably acts as an activator of the anaphase promoting complex/cyclosome (APC/C) that promotes the ubiquitin ligase activity and substrate specificity of the APC/C (By similarity).
Female meiosis-specific activator of the anaphase promoting complex/cyclosome (APC/C) (PubMed:17251266, PubMed:18020708). Required for the completion of meiosis in oocytes (PubMed:11252055, PubMed:10683177, PubMed:10924478, PubMed:17251266, PubMed:18020708). Activates the ubiquitin ligase activity and substrate specificity of APC/C and triggers the sequential degradation of mitotic cyclins in meiosis (PubMed:17251266, PubMed:18020708). Promotes the ubiquitination and degradation of CycA early in meiosis I and the degradation of CycB and CycB3 after egg activation (PubMed:18020708). Promotes degradation of mtrm at the oocyte-to-embryo transition (PubMed:24019759).
Controls wing pigmentation patterning, possibly by regulating scale cell development (PubMed:27251284). Probably acts as an activator of the anaphase promoting complex/cyclosome (APC/C) that promotes the ubiquitin ligase activity and substrate specificity of the APC/C (By similarity).
Female meiosis-specific activator of the anaphase promoting complex/cyclosome (APC/C) (PubMed:17251266, PubMed:18020708). Required for the completion of meiosis in oocytes (PubMed:11252055, PubMed:10683177, PubMed:10924478, PubMed:17251266, PubMed:18020708). Activates the ubiquitin ligase activity and substrate specificity of APC/C and triggers the sequential degradation of mitotic cyclins in meiosis (PubMed:17251266, PubMed:18020708). Promotes the ubiquitination and degradation of CycA early in meiosis I and the degradation of CycB and CycB3 after egg activation (PubMed:18020708). Promotes degradation of mtrm at the oocyte-to-embryo transition (PubMed:24019759).
Subunit
Interacts with anaphase promoting complex/cyclosome (APC/C) subunits Cdc16 and Cdc27 (PubMed:18020708). Interacts with mtrm (PubMed:24019759).
Similarity
Belongs to the WD repeat CORT family.
Keywords
Cytoplasm
Repeat
WD repeat
Alternative splicing
Complete proteome
Developmental protein
Differentiation
Meiosis
Oogenesis
Reference proteome
Ubl conjugation
Feature
chain Protein cortex
splice variant In isoform B4.
splice variant In isoform B4.
Uniprot
A0A212EI88
L7X1N7
P0DOB9
A0A2A4J7B4
C9S286
P0DOB8
+ More
A0A3S2L785 A0A2H1VEY5 D0ABB1 P0DOC0-5 P0DOC0-7 A0A173FD67 A0A194QXL3 A0A173FD09 P0DOC0 S4PYC8 A0A194QGP3 P0DOC0-2 P0DOC0-4 P0DOC0-9 P0DOC0-3 P0DOC0-10 A0A067QYG4 A0A2J7RLQ2 A0A1B6CX61 B4LRF9 W8BYQ2 A0A0Q9XE59 B4KJU8 A0A1B6MRK4 A0A1B6E7D5 A0A034W8P7 A0A0A1X220 A0A0R3NV72 Q29JW4 B4GJ14 A0A0L0BXI1 A0A1I8MQ76 A0A0K8V4F0 A0A1B6J1A1 A0A0K8VDK2 A0A0K8WJC8 A0A0Q5WP77 B3N5V3 K7JWH4 E0VHE7 A0A3B0K7B4 A0A291S6V3 B4JPS1 A0A0C9PZK9 A0A0C9RF39 A0A0R1DJR2 B4P098 D6WE73 A0A224XNL6 B4HXW0 M9PCR0 Q960N3 B4Q503 A0A232ELS4 B4MZ25 A0A0C9PMR9 B0XET5 E0S6S7 F2TXX8 A0A1B0CWH2 A0A087ZTL1 A0A1X7VQX7 A0A1W4UBE6 V8PF70 A0A1Q3EVD3 A0A1W4WIS2 W5JVQ7 A0A1B0GQP2 A0A1W4WUW4 S7PBU5 B0WYP0 L5LYF6 F6UE96 A0A1A8S7Z6 A0A2A3E4C6 A0A182F654 W5JAB6 A0A1A8MTQ3 A0A3B3S620 Q8AVG7 Q6P867 A0A2P6L0U5 A0A151P6B9 M7C196 A0A2G8K7L9 A0A1U8D6L4 T1J1W4 A0A2C9JHF3 K9TNH7 K1R5H7
A0A3S2L785 A0A2H1VEY5 D0ABB1 P0DOC0-5 P0DOC0-7 A0A173FD67 A0A194QXL3 A0A173FD09 P0DOC0 S4PYC8 A0A194QGP3 P0DOC0-2 P0DOC0-4 P0DOC0-9 P0DOC0-3 P0DOC0-10 A0A067QYG4 A0A2J7RLQ2 A0A1B6CX61 B4LRF9 W8BYQ2 A0A0Q9XE59 B4KJU8 A0A1B6MRK4 A0A1B6E7D5 A0A034W8P7 A0A0A1X220 A0A0R3NV72 Q29JW4 B4GJ14 A0A0L0BXI1 A0A1I8MQ76 A0A0K8V4F0 A0A1B6J1A1 A0A0K8VDK2 A0A0K8WJC8 A0A0Q5WP77 B3N5V3 K7JWH4 E0VHE7 A0A3B0K7B4 A0A291S6V3 B4JPS1 A0A0C9PZK9 A0A0C9RF39 A0A0R1DJR2 B4P098 D6WE73 A0A224XNL6 B4HXW0 M9PCR0 Q960N3 B4Q503 A0A232ELS4 B4MZ25 A0A0C9PMR9 B0XET5 E0S6S7 F2TXX8 A0A1B0CWH2 A0A087ZTL1 A0A1X7VQX7 A0A1W4UBE6 V8PF70 A0A1Q3EVD3 A0A1W4WIS2 W5JVQ7 A0A1B0GQP2 A0A1W4WUW4 S7PBU5 B0WYP0 L5LYF6 F6UE96 A0A1A8S7Z6 A0A2A3E4C6 A0A182F654 W5JAB6 A0A1A8MTQ3 A0A3B3S620 Q8AVG7 Q6P867 A0A2P6L0U5 A0A151P6B9 M7C196 A0A2G8K7L9 A0A1U8D6L4 T1J1W4 A0A2C9JHF3 K9TNH7 K1R5H7
Pubmed
22118469
23674305
27251285
27251284
26354079
23622113
+ More
24845553 17994087 24495485 25348373 25830018 15632085 26108605 25315136 20075255 20566863 28992199 17550304 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11252055 12537569 10683177 10924478 17251266 18020708 24019759 22936249 28648823 20865802 24297900 20920257 23761445 20431018 29240929 27762356 22293439 23624526 29023486 15562597 22992520
24845553 17994087 24495485 25348373 25830018 15632085 26108605 25315136 20075255 20566863 28992199 17550304 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 11252055 12537569 10683177 10924478 17251266 18020708 24019759 22936249 28648823 20865802 24297900 20920257 23761445 20431018 29240929 27762356 22293439 23624526 29023486 15562597 22992520
EMBL
AGBW02014684
OWR41180.1
KC469893
AGC92688.2
NWSH01002612
PCG67851.1
+ More
CU463862 CBH09266.1 RSAL01000108 RVE47260.1 ODYU01002201 SOQ39387.1 FP236845 CBH09306.1 KT235895 KT235896 KT235897 KT235898 KT235899 KT235900 KT235901 KT235902 KT235903 KT235904 KT235905 KT235906 KT182637 ANG83461.1 KQ461155 KPJ08316.1 ANG83460.1 GAIX01003513 JAA89047.1 KQ459232 KPJ02616.1 KK852878 KDR14508.1 NEVH01002684 PNF41761.1 GEDC01019219 JAS18079.1 CH940649 EDW64629.2 GAMC01000040 JAC06516.1 CH933807 KRG03284.1 EDW12551.2 GEBQ01001407 JAT38570.1 GEDC01003534 JAS33764.1 GAKP01007873 JAC51079.1 GBXI01008923 JAD05369.1 CH379062 KRT04999.1 EAL32847.2 CH479184 EDW37328.1 JRES01001184 KNC24701.1 GDHF01018522 JAI33792.1 GECU01014770 JAS92936.1 GDHF01015689 JAI36625.1 GDHF01001135 JAI51179.1 CH954177 KQS70632.1 EDV59112.1 AAZX01000605 AAZX01018649 DS235169 EEB12803.1 OUUW01000006 SPP81536.1 MF432985 ATL75336.1 CH916372 EDV98901.1 GBYB01006993 JAG76760.1 GBYB01007005 JAG76772.1 CM000157 KRJ97455.1 EDW87856.2 KQ971324 EFA01168.2 GFTR01006847 JAW09579.1 CH480818 EDW51890.1 AE014134 AGB92680.1 AY033478 AY051966 CM000361 CM002910 EDX03995.1 KMY88575.1 NNAY01003513 OXU19297.1 CH963913 EDW77421.1 GBYB01002428 JAG72195.1 DS232868 EDS26217.1 CP001945 ADM11412.1 GL832956 EGD76237.1 AJWK01032468 AZIM01000193 ETE72658.1 GFDL01015809 JAV19236.1 ADMH02000329 ETN66934.1 AJVK01017759 KE162280 EPQ07678.1 DS232195 EDS37150.1 KB106370 ELK31141.1 AAMC01002118 HAEH01022295 SBS13723.1 KZ288419 PBC26009.1 ADMH02001847 ETN60906.1 HAEF01018863 SBR60022.1 BC042288 CM004473 AAH42288.1 OCT82635.1 BC061363 CR760268 AAH61363.1 CAJ83291.1 MWRG01002831 PRD32199.1 AKHW03000704 KYO44594.1 KB533688 EMP34162.1 MRZV01000808 PIK44016.1 JH431790 CP003607 AFY83701.1 JH816764 EKC38794.1
CU463862 CBH09266.1 RSAL01000108 RVE47260.1 ODYU01002201 SOQ39387.1 FP236845 CBH09306.1 KT235895 KT235896 KT235897 KT235898 KT235899 KT235900 KT235901 KT235902 KT235903 KT235904 KT235905 KT235906 KT182637 ANG83461.1 KQ461155 KPJ08316.1 ANG83460.1 GAIX01003513 JAA89047.1 KQ459232 KPJ02616.1 KK852878 KDR14508.1 NEVH01002684 PNF41761.1 GEDC01019219 JAS18079.1 CH940649 EDW64629.2 GAMC01000040 JAC06516.1 CH933807 KRG03284.1 EDW12551.2 GEBQ01001407 JAT38570.1 GEDC01003534 JAS33764.1 GAKP01007873 JAC51079.1 GBXI01008923 JAD05369.1 CH379062 KRT04999.1 EAL32847.2 CH479184 EDW37328.1 JRES01001184 KNC24701.1 GDHF01018522 JAI33792.1 GECU01014770 JAS92936.1 GDHF01015689 JAI36625.1 GDHF01001135 JAI51179.1 CH954177 KQS70632.1 EDV59112.1 AAZX01000605 AAZX01018649 DS235169 EEB12803.1 OUUW01000006 SPP81536.1 MF432985 ATL75336.1 CH916372 EDV98901.1 GBYB01006993 JAG76760.1 GBYB01007005 JAG76772.1 CM000157 KRJ97455.1 EDW87856.2 KQ971324 EFA01168.2 GFTR01006847 JAW09579.1 CH480818 EDW51890.1 AE014134 AGB92680.1 AY033478 AY051966 CM000361 CM002910 EDX03995.1 KMY88575.1 NNAY01003513 OXU19297.1 CH963913 EDW77421.1 GBYB01002428 JAG72195.1 DS232868 EDS26217.1 CP001945 ADM11412.1 GL832956 EGD76237.1 AJWK01032468 AZIM01000193 ETE72658.1 GFDL01015809 JAV19236.1 ADMH02000329 ETN66934.1 AJVK01017759 KE162280 EPQ07678.1 DS232195 EDS37150.1 KB106370 ELK31141.1 AAMC01002118 HAEH01022295 SBS13723.1 KZ288419 PBC26009.1 ADMH02001847 ETN60906.1 HAEF01018863 SBR60022.1 BC042288 CM004473 AAH42288.1 OCT82635.1 BC061363 CR760268 AAH61363.1 CAJ83291.1 MWRG01002831 PRD32199.1 AKHW03000704 KYO44594.1 KB533688 EMP34162.1 MRZV01000808 PIK44016.1 JH431790 CP003607 AFY83701.1 JH816764 EKC38794.1
Proteomes
UP000007151
UP000218220
UP000283053
UP000053240
UP000053268
UP000027135
+ More
UP000235965 UP000008792 UP000009192 UP000001819 UP000008744 UP000037069 UP000095301 UP000008711 UP000002358 UP000009046 UP000268350 UP000001070 UP000002282 UP000007266 UP000001292 UP000000803 UP000000304 UP000215335 UP000007798 UP000002320 UP000002313 UP000007799 UP000092461 UP000005203 UP000007879 UP000192221 UP000192223 UP000000673 UP000092462 UP000008143 UP000242457 UP000069272 UP000261540 UP000186698 UP000050525 UP000031443 UP000230750 UP000189705 UP000076420 UP000010367 UP000005408
UP000235965 UP000008792 UP000009192 UP000001819 UP000008744 UP000037069 UP000095301 UP000008711 UP000002358 UP000009046 UP000268350 UP000001070 UP000002282 UP000007266 UP000001292 UP000000803 UP000000304 UP000215335 UP000007798 UP000002320 UP000002313 UP000007799 UP000092461 UP000005203 UP000007879 UP000192221 UP000192223 UP000000673 UP000092462 UP000008143 UP000242457 UP000069272 UP000261540 UP000186698 UP000050525 UP000031443 UP000230750 UP000189705 UP000076420 UP000010367 UP000005408
Interpro
Gene 3D
ProteinModelPortal
A0A212EI88
L7X1N7
P0DOB9
A0A2A4J7B4
C9S286
P0DOB8
+ More
A0A3S2L785 A0A2H1VEY5 D0ABB1 P0DOC0-5 P0DOC0-7 A0A173FD67 A0A194QXL3 A0A173FD09 P0DOC0 S4PYC8 A0A194QGP3 P0DOC0-2 P0DOC0-4 P0DOC0-9 P0DOC0-3 P0DOC0-10 A0A067QYG4 A0A2J7RLQ2 A0A1B6CX61 B4LRF9 W8BYQ2 A0A0Q9XE59 B4KJU8 A0A1B6MRK4 A0A1B6E7D5 A0A034W8P7 A0A0A1X220 A0A0R3NV72 Q29JW4 B4GJ14 A0A0L0BXI1 A0A1I8MQ76 A0A0K8V4F0 A0A1B6J1A1 A0A0K8VDK2 A0A0K8WJC8 A0A0Q5WP77 B3N5V3 K7JWH4 E0VHE7 A0A3B0K7B4 A0A291S6V3 B4JPS1 A0A0C9PZK9 A0A0C9RF39 A0A0R1DJR2 B4P098 D6WE73 A0A224XNL6 B4HXW0 M9PCR0 Q960N3 B4Q503 A0A232ELS4 B4MZ25 A0A0C9PMR9 B0XET5 E0S6S7 F2TXX8 A0A1B0CWH2 A0A087ZTL1 A0A1X7VQX7 A0A1W4UBE6 V8PF70 A0A1Q3EVD3 A0A1W4WIS2 W5JVQ7 A0A1B0GQP2 A0A1W4WUW4 S7PBU5 B0WYP0 L5LYF6 F6UE96 A0A1A8S7Z6 A0A2A3E4C6 A0A182F654 W5JAB6 A0A1A8MTQ3 A0A3B3S620 Q8AVG7 Q6P867 A0A2P6L0U5 A0A151P6B9 M7C196 A0A2G8K7L9 A0A1U8D6L4 T1J1W4 A0A2C9JHF3 K9TNH7 K1R5H7
A0A3S2L785 A0A2H1VEY5 D0ABB1 P0DOC0-5 P0DOC0-7 A0A173FD67 A0A194QXL3 A0A173FD09 P0DOC0 S4PYC8 A0A194QGP3 P0DOC0-2 P0DOC0-4 P0DOC0-9 P0DOC0-3 P0DOC0-10 A0A067QYG4 A0A2J7RLQ2 A0A1B6CX61 B4LRF9 W8BYQ2 A0A0Q9XE59 B4KJU8 A0A1B6MRK4 A0A1B6E7D5 A0A034W8P7 A0A0A1X220 A0A0R3NV72 Q29JW4 B4GJ14 A0A0L0BXI1 A0A1I8MQ76 A0A0K8V4F0 A0A1B6J1A1 A0A0K8VDK2 A0A0K8WJC8 A0A0Q5WP77 B3N5V3 K7JWH4 E0VHE7 A0A3B0K7B4 A0A291S6V3 B4JPS1 A0A0C9PZK9 A0A0C9RF39 A0A0R1DJR2 B4P098 D6WE73 A0A224XNL6 B4HXW0 M9PCR0 Q960N3 B4Q503 A0A232ELS4 B4MZ25 A0A0C9PMR9 B0XET5 E0S6S7 F2TXX8 A0A1B0CWH2 A0A087ZTL1 A0A1X7VQX7 A0A1W4UBE6 V8PF70 A0A1Q3EVD3 A0A1W4WIS2 W5JVQ7 A0A1B0GQP2 A0A1W4WUW4 S7PBU5 B0WYP0 L5LYF6 F6UE96 A0A1A8S7Z6 A0A2A3E4C6 A0A182F654 W5JAB6 A0A1A8MTQ3 A0A3B3S620 Q8AVG7 Q6P867 A0A2P6L0U5 A0A151P6B9 M7C196 A0A2G8K7L9 A0A1U8D6L4 T1J1W4 A0A2C9JHF3 K9TNH7 K1R5H7
PDB
6F0X
E-value=1.22751e-10,
Score=159
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
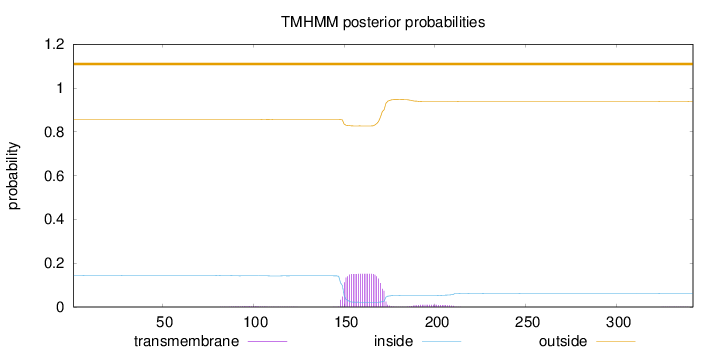
Length:
342
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.59169
Exp number, first 60 AAs:
0.00114
Total prob of N-in:
0.14467
outside
1 - 342
Population Genetic Test Statistics
Pi
193.829021
Theta
171.845889
Tajima's D
0.254543
CLR
0.12611
CSRT
0.44212789360532
Interpretation
Uncertain
