Pre Gene Modal
BGIBMGA005557
Annotation
ATP_binding_protein_[Bombyx_mori]
Full name
Cancer-related nucleoside-triphosphatase homolog
+ More
Cancer-related nucleoside-triphosphatase
Cancer-related nucleoside-triphosphatase
Alternative Name
Nucleoside triphosphate phosphohydrolase
Location in the cell
Mitochondrial Reliability : 1.143 Nuclear Reliability : 1.047
Sequence
CDS
ATGTCTAAAAATCCTAATACAACAATAAGGTTTTTTATATTAACAGGCGATCCTGGCATCGGTAAAACTACTCTGACCAAGAAAATATCTTCGCTCTTAGAAGCAGAAGGTGTAAAAGTTAATGGTTTTATTACTGAAGAAGTCAGAAATAAAGGAGTAAGAGAAGGCTTCGATGTGGTTACTAGTGCAGGAGTCAGGGGCCGTTTGGCTAGGGACCAAGATCTCATCACTATCCCTATTAAGCGCAAAATTGGTAAATACGGGGTTTTTATTGAAGAGTTTGAAAATATTGCATTGCCATGTTTAAACGAGACAACATCACAACCAAACTTACTAGTAATAGATGAAATTGGAAGCATGGAATTATCTAGTAAAAAGTTTAAAAATGTAATCGAAAATATTTTTAGTTCTGAGCAAAATTCTTACAATGTTGTGTTGGCAACCATACCTGTTAAAAACAGGTCTGCTCTTGTGGAGAACATTAGAAGTCATCCTAAAGCTAAAGTTTGGGTGATTACAAAAAAGAATAGAAATAAACTACATGAAGAAGTTATAAACGAAATGAAATCTGTTTTGAGTTTGATTTAA
Protein
MSKNPNTTIRFFILTGDPGIGKTTLTKKISSLLEAEGVKVNGFITEEVRNKGVREGFDVVTSAGVRGRLARDQDLITIPIKRKIGKYGVFIEEFENIALPCLNETTSQPNLLVIDEIGSMELSSKKFKNVIENIFSSEQNSYNVVLATIPVKNRSALVENIRSHPKAKVWVITKKNRNKLHEEVINEMKSVLSLI
Summary
Description
Has nucleotide phosphatase activity towards ATP, GTP, CTP, TTP and UTP. Hydrolyzes nucleoside diphosphates with lower efficiency (By similarity).
Has nucleotide phosphatase activity towards ATP, GTP, CTP, TTP and UTP. Hydrolyzes nucleoside diphosphates with lower efficiency.
Has nucleotide phosphatase activity towards ATP, GTP, CTP, TTP and UTP. Hydrolyzes nucleoside diphosphates with lower efficiency.
Catalytic Activity
a ribonucleoside 5'-triphosphate + H2O = a ribonucleoside 5'-diphosphate + H(+) + phosphate
Biophysicochemical Properties
5 uM for ATP
46 uM for ADP
4 uM for GTP
6 uM for CTP
7 uM for TTP
46 uM for ADP
4 uM for GTP
6 uM for CTP
7 uM for TTP
Subunit
Monomer.
Similarity
Belongs to the THEP1 NTPase family.
Keywords
Acetylation
ATP-binding
Complete proteome
Hydrolase
Nucleotide-binding
Reference proteome
3D-structure
Polymorphism
Feature
chain Cancer-related nucleoside-triphosphatase homolog
sequence variant In dbSNP:rs12123482.
sequence variant In dbSNP:rs12123482.
Uniprot
Q2F631
H9J7R5
A0A173FDM5
A0A2H1VHW6
A0A1E1W850
L7X3V9
+ More
A0A194QC99 A0A212EI73 A0A1E1W9D0 A0A0L7L2L0 A0A194QS57 A0A2A4IUN2 B4QJM2 A0A1A9YTA1 B4IAJ2 A0A1A9VIS2 Q4V5T6 Q9VP84 B4PE79 B4J1L3 L7M6F5 K1Q1T3 A0A0P6K5D2 A0A1Z5L7L0 A0A0L0BWL5 B3NIV2 B4ITI0 A0A091DQ47 Q2M163 F1MM52 W5NZG4 H0VCP2 Q1LZ78 A0A1W4UXP8 A0A3B0J4U7 A0A2Y9DEX4 F6W380 A0A1I8M1G6 A0A131XH92 G3GWZ6 A0A2K5ND48 A0A2K6CY20 A0A2K5TSZ0 A0A2K5YE68 A0A0D9RCU8 A0A096MSK8 H9ZEC8 K9IYN6 A0A2D0QV73 I3MW13 B4LES7 A0A1Y1K683 G1T786 A0A340Y5P8 H2N3D9 A0A2U4BVF2 B3M6U6 A0A2C9LU97 A0A383ZFF4 A0A2R9AG00 K7CPD8 Q5TDE9 Q9BSD7 A0A1S3WBR3 G1Q846 A0A1U7QEM7 H0WQB7 G3S2M6 A0A3Q7VFU1 A0A2Y9LU58 A0A2I3HB03 A0A3Q2CBY7 A0A341AVD2 A0A3Q7TGB0 A0A2J7R6P6 A0A2K5R493 V3ZFT9 A0A3B4CS53 A0A3Q3GDA7 A0A1J1IQ59 A0A0M3QWA2 A0A2K5BV08 A0A023GF21 G1K8P1 A0A2K6T8C7 A0A2Y9FP34 L8IVF3 A0A023FIW2 A0A337S2Z4 H3D7D3 A0A0P4WBG2 A0A3Q1IGF5 A0A2U3WIR0 G3MLY6 B4KVY7 A0A1S3JTY1 A0A0K8RYF7 G3TP10 F1RGU8 C3KJN0
A0A194QC99 A0A212EI73 A0A1E1W9D0 A0A0L7L2L0 A0A194QS57 A0A2A4IUN2 B4QJM2 A0A1A9YTA1 B4IAJ2 A0A1A9VIS2 Q4V5T6 Q9VP84 B4PE79 B4J1L3 L7M6F5 K1Q1T3 A0A0P6K5D2 A0A1Z5L7L0 A0A0L0BWL5 B3NIV2 B4ITI0 A0A091DQ47 Q2M163 F1MM52 W5NZG4 H0VCP2 Q1LZ78 A0A1W4UXP8 A0A3B0J4U7 A0A2Y9DEX4 F6W380 A0A1I8M1G6 A0A131XH92 G3GWZ6 A0A2K5ND48 A0A2K6CY20 A0A2K5TSZ0 A0A2K5YE68 A0A0D9RCU8 A0A096MSK8 H9ZEC8 K9IYN6 A0A2D0QV73 I3MW13 B4LES7 A0A1Y1K683 G1T786 A0A340Y5P8 H2N3D9 A0A2U4BVF2 B3M6U6 A0A2C9LU97 A0A383ZFF4 A0A2R9AG00 K7CPD8 Q5TDE9 Q9BSD7 A0A1S3WBR3 G1Q846 A0A1U7QEM7 H0WQB7 G3S2M6 A0A3Q7VFU1 A0A2Y9LU58 A0A2I3HB03 A0A3Q2CBY7 A0A341AVD2 A0A3Q7TGB0 A0A2J7R6P6 A0A2K5R493 V3ZFT9 A0A3B4CS53 A0A3Q3GDA7 A0A1J1IQ59 A0A0M3QWA2 A0A2K5BV08 A0A023GF21 G1K8P1 A0A2K6T8C7 A0A2Y9FP34 L8IVF3 A0A023FIW2 A0A337S2Z4 H3D7D3 A0A0P4WBG2 A0A3Q1IGF5 A0A2U3WIR0 G3MLY6 B4KVY7 A0A1S3JTY1 A0A0K8RYF7 G3TP10 F1RGU8 C3KJN0
EC Number
3.6.1.15
Pubmed
19121390
27251284
23674305
26354079
22118469
26227816
+ More
17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25576852 22992520 28528879 26108605 15632085 19393038 20809919 21993624 19892987 25315136 28049606 21804562 25319552 18057021 28004739 15562597 22722832 16136131 11181995 15489334 19413330 19608861 21269460 25944712 17291528 23254933 22751099 17975172 15496914 22216098 25727380 30723633
17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25576852 22992520 28528879 26108605 15632085 19393038 20809919 21993624 19892987 25315136 28049606 21804562 25319552 18057021 28004739 15562597 22722832 16136131 11181995 15489334 19413330 19608861 21269460 25944712 17291528 23254933 22751099 17975172 15496914 22216098 25727380 30723633
EMBL
DQ311241
ABD36186.1
BABH01020258
BABH01020259
KT182637
ANG83454.1
+ More
ODYU01002629 SOQ40386.1 GDQN01007929 JAT83125.1 KC469893 AGC92685.1 KQ459232 KPJ02610.1 AGBW02014684 OWR41186.1 GDQN01007478 JAT83576.1 JTDY01003352 KOB69687.1 KQ461155 KPJ08322.1 NWSH01007700 PCG62850.1 CM000363 CM002912 EDX11322.1 KMZ00919.1 CH480826 EDW44305.1 BT022570 AAY54986.1 AE014296 BT022513 AAF51671.2 AAY54929.1 CM000159 EDW95092.1 CH916366 EDV96933.1 GACK01006355 JAA58679.1 JH816081 EKC25319.1 GEBF01003302 JAO00331.1 GFJQ02003695 JAW03275.1 JRES01001238 KNC24423.1 CH954178 EDV52598.1 CH891700 EDW99693.1 KN122008 KFO34234.1 CH379069 EAL30712.3 AMGL01070647 AAKN02021646 AAKN02021647 BC116158 OUUW01000002 SPP76904.1 GEFH01002674 JAP65907.1 JH000057 EGW03340.1 AQIA01000466 AQIB01131522 AHZZ02017912 AHZZ02017913 JU336699 JU477490 AFE80452.1 AFH34294.1 GABZ01008101 JAA45424.1 AGTP01017295 AGTP01017296 CH940647 EDW70184.1 KRF84753.1 GEZM01091334 JAV56903.1 AAGW02026259 ABGA01149665 ABGA01149666 ABGA01149667 NDHI03003462 PNJ43528.1 CH902618 EDV39782.1 AJFE02086383 AACZ04026634 GABC01010870 GABF01010765 GABD01004916 GABE01002243 NBAG03000266 JAA00468.1 JAA11380.1 JAA28184.1 JAA42496.1 PNI55179.1 AK316591 CH471098 BAG38178.1 EAW69979.1 AF416713 BC005102 AAPE02053529 AAPE02053530 AAQR03135511 CABD030009447 ADFV01036771 ADFV01036772 NEVH01006756 PNF36508.1 KB203827 ESO82982.1 CVRI01000057 CRL01866.1 CP012525 ALC43793.1 GBBM01002727 JAC32691.1 JH880752 ELR59137.1 GBBK01003909 JAC20573.1 AANG04003799 GDRN01055236 JAI66001.1 JO842887 AEO34504.1 CH933809 EDW19538.1 GBKC01000580 JAG45490.1 AEMK02000096 DQIR01231014 HDB86491.1 BT083145 ACQ58852.1
ODYU01002629 SOQ40386.1 GDQN01007929 JAT83125.1 KC469893 AGC92685.1 KQ459232 KPJ02610.1 AGBW02014684 OWR41186.1 GDQN01007478 JAT83576.1 JTDY01003352 KOB69687.1 KQ461155 KPJ08322.1 NWSH01007700 PCG62850.1 CM000363 CM002912 EDX11322.1 KMZ00919.1 CH480826 EDW44305.1 BT022570 AAY54986.1 AE014296 BT022513 AAF51671.2 AAY54929.1 CM000159 EDW95092.1 CH916366 EDV96933.1 GACK01006355 JAA58679.1 JH816081 EKC25319.1 GEBF01003302 JAO00331.1 GFJQ02003695 JAW03275.1 JRES01001238 KNC24423.1 CH954178 EDV52598.1 CH891700 EDW99693.1 KN122008 KFO34234.1 CH379069 EAL30712.3 AMGL01070647 AAKN02021646 AAKN02021647 BC116158 OUUW01000002 SPP76904.1 GEFH01002674 JAP65907.1 JH000057 EGW03340.1 AQIA01000466 AQIB01131522 AHZZ02017912 AHZZ02017913 JU336699 JU477490 AFE80452.1 AFH34294.1 GABZ01008101 JAA45424.1 AGTP01017295 AGTP01017296 CH940647 EDW70184.1 KRF84753.1 GEZM01091334 JAV56903.1 AAGW02026259 ABGA01149665 ABGA01149666 ABGA01149667 NDHI03003462 PNJ43528.1 CH902618 EDV39782.1 AJFE02086383 AACZ04026634 GABC01010870 GABF01010765 GABD01004916 GABE01002243 NBAG03000266 JAA00468.1 JAA11380.1 JAA28184.1 JAA42496.1 PNI55179.1 AK316591 CH471098 BAG38178.1 EAW69979.1 AF416713 BC005102 AAPE02053529 AAPE02053530 AAQR03135511 CABD030009447 ADFV01036771 ADFV01036772 NEVH01006756 PNF36508.1 KB203827 ESO82982.1 CVRI01000057 CRL01866.1 CP012525 ALC43793.1 GBBM01002727 JAC32691.1 JH880752 ELR59137.1 GBBK01003909 JAC20573.1 AANG04003799 GDRN01055236 JAI66001.1 JO842887 AEO34504.1 CH933809 EDW19538.1 GBKC01000580 JAG45490.1 AEMK02000096 DQIR01231014 HDB86491.1 BT083145 ACQ58852.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000037510
UP000053240
UP000218220
+ More
UP000000304 UP000092443 UP000001292 UP000078200 UP000000803 UP000002282 UP000001070 UP000005408 UP000037069 UP000008711 UP000028990 UP000001819 UP000009136 UP000002356 UP000005447 UP000192221 UP000268350 UP000248480 UP000002281 UP000095301 UP000001075 UP000233060 UP000233120 UP000233100 UP000233140 UP000029965 UP000028761 UP000221080 UP000005215 UP000008792 UP000001811 UP000265300 UP000001595 UP000245320 UP000007801 UP000076420 UP000261681 UP000240080 UP000002277 UP000005640 UP000079721 UP000001074 UP000189706 UP000005225 UP000001519 UP000286642 UP000248483 UP000001073 UP000265020 UP000252040 UP000286640 UP000235965 UP000233040 UP000030746 UP000261440 UP000261660 UP000183832 UP000092553 UP000233020 UP000001646 UP000233220 UP000248484 UP000011712 UP000007303 UP000265040 UP000245340 UP000009192 UP000085678 UP000007646 UP000008227
UP000000304 UP000092443 UP000001292 UP000078200 UP000000803 UP000002282 UP000001070 UP000005408 UP000037069 UP000008711 UP000028990 UP000001819 UP000009136 UP000002356 UP000005447 UP000192221 UP000268350 UP000248480 UP000002281 UP000095301 UP000001075 UP000233060 UP000233120 UP000233100 UP000233140 UP000029965 UP000028761 UP000221080 UP000005215 UP000008792 UP000001811 UP000265300 UP000001595 UP000245320 UP000007801 UP000076420 UP000261681 UP000240080 UP000002277 UP000005640 UP000079721 UP000001074 UP000189706 UP000005225 UP000001519 UP000286642 UP000248483 UP000001073 UP000265020 UP000252040 UP000286640 UP000235965 UP000233040 UP000030746 UP000261440 UP000261660 UP000183832 UP000092553 UP000233020 UP000001646 UP000233220 UP000248484 UP000011712 UP000007303 UP000265040 UP000245340 UP000009192 UP000085678 UP000007646 UP000008227
Pfam
PF03266 NTPase_1
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
Q2F631
H9J7R5
A0A173FDM5
A0A2H1VHW6
A0A1E1W850
L7X3V9
+ More
A0A194QC99 A0A212EI73 A0A1E1W9D0 A0A0L7L2L0 A0A194QS57 A0A2A4IUN2 B4QJM2 A0A1A9YTA1 B4IAJ2 A0A1A9VIS2 Q4V5T6 Q9VP84 B4PE79 B4J1L3 L7M6F5 K1Q1T3 A0A0P6K5D2 A0A1Z5L7L0 A0A0L0BWL5 B3NIV2 B4ITI0 A0A091DQ47 Q2M163 F1MM52 W5NZG4 H0VCP2 Q1LZ78 A0A1W4UXP8 A0A3B0J4U7 A0A2Y9DEX4 F6W380 A0A1I8M1G6 A0A131XH92 G3GWZ6 A0A2K5ND48 A0A2K6CY20 A0A2K5TSZ0 A0A2K5YE68 A0A0D9RCU8 A0A096MSK8 H9ZEC8 K9IYN6 A0A2D0QV73 I3MW13 B4LES7 A0A1Y1K683 G1T786 A0A340Y5P8 H2N3D9 A0A2U4BVF2 B3M6U6 A0A2C9LU97 A0A383ZFF4 A0A2R9AG00 K7CPD8 Q5TDE9 Q9BSD7 A0A1S3WBR3 G1Q846 A0A1U7QEM7 H0WQB7 G3S2M6 A0A3Q7VFU1 A0A2Y9LU58 A0A2I3HB03 A0A3Q2CBY7 A0A341AVD2 A0A3Q7TGB0 A0A2J7R6P6 A0A2K5R493 V3ZFT9 A0A3B4CS53 A0A3Q3GDA7 A0A1J1IQ59 A0A0M3QWA2 A0A2K5BV08 A0A023GF21 G1K8P1 A0A2K6T8C7 A0A2Y9FP34 L8IVF3 A0A023FIW2 A0A337S2Z4 H3D7D3 A0A0P4WBG2 A0A3Q1IGF5 A0A2U3WIR0 G3MLY6 B4KVY7 A0A1S3JTY1 A0A0K8RYF7 G3TP10 F1RGU8 C3KJN0
A0A194QC99 A0A212EI73 A0A1E1W9D0 A0A0L7L2L0 A0A194QS57 A0A2A4IUN2 B4QJM2 A0A1A9YTA1 B4IAJ2 A0A1A9VIS2 Q4V5T6 Q9VP84 B4PE79 B4J1L3 L7M6F5 K1Q1T3 A0A0P6K5D2 A0A1Z5L7L0 A0A0L0BWL5 B3NIV2 B4ITI0 A0A091DQ47 Q2M163 F1MM52 W5NZG4 H0VCP2 Q1LZ78 A0A1W4UXP8 A0A3B0J4U7 A0A2Y9DEX4 F6W380 A0A1I8M1G6 A0A131XH92 G3GWZ6 A0A2K5ND48 A0A2K6CY20 A0A2K5TSZ0 A0A2K5YE68 A0A0D9RCU8 A0A096MSK8 H9ZEC8 K9IYN6 A0A2D0QV73 I3MW13 B4LES7 A0A1Y1K683 G1T786 A0A340Y5P8 H2N3D9 A0A2U4BVF2 B3M6U6 A0A2C9LU97 A0A383ZFF4 A0A2R9AG00 K7CPD8 Q5TDE9 Q9BSD7 A0A1S3WBR3 G1Q846 A0A1U7QEM7 H0WQB7 G3S2M6 A0A3Q7VFU1 A0A2Y9LU58 A0A2I3HB03 A0A3Q2CBY7 A0A341AVD2 A0A3Q7TGB0 A0A2J7R6P6 A0A2K5R493 V3ZFT9 A0A3B4CS53 A0A3Q3GDA7 A0A1J1IQ59 A0A0M3QWA2 A0A2K5BV08 A0A023GF21 G1K8P1 A0A2K6T8C7 A0A2Y9FP34 L8IVF3 A0A023FIW2 A0A337S2Z4 H3D7D3 A0A0P4WBG2 A0A3Q1IGF5 A0A2U3WIR0 G3MLY6 B4KVY7 A0A1S3JTY1 A0A0K8RYF7 G3TP10 F1RGU8 C3KJN0
PDB
2I3B
E-value=7.19376e-29,
Score=313
Ontologies
PATHWAY
Topology
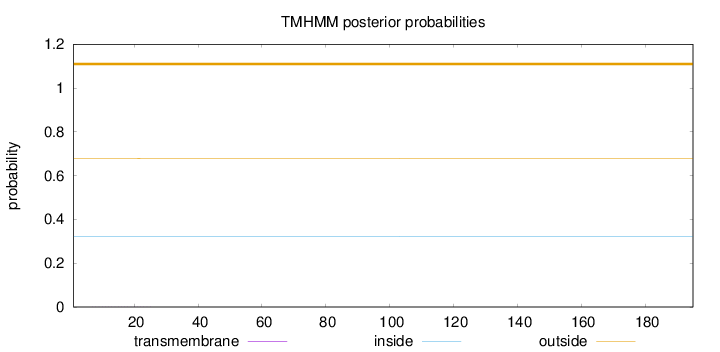
Length:
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00414
Exp number, first 60 AAs:
0.00357
Total prob of N-in:
0.32215
outside
1 - 195
Population Genetic Test Statistics
Pi
128.581457
Theta
148.834265
Tajima's D
-0.295655
CLR
258.648726
CSRT
0.293035348232588
Interpretation
Uncertain
