Gene
KWMTBOMO10111
Pre Gene Modal
BGIBMGA005555
Annotation
PREDICTED:_acid_beta-fructofuranosidase-like_[Bombyx_mori]
Full name
Sucrose-6-phosphate hydrolase
Alternative Name
Invertase
Location in the cell
Extracellular Reliability : 1.111
Sequence
CDS
ATGGACGTTTTAGTTGTGGCATTATATTTGCGTTTCGTTTGCGCGGTCTTCGCGGTAGACTTGTCTCATATTGAGACTTTTGTACAGGAGAATCGTTACTGTTTGAAACAGAGATATCGTCCACTGTACCACATATCAGCGGCCCACGCATGGACGAACCATCCCAGCGCGTTCGTGTATTACAAACGGCAGTACCATATCTTTTACCAATATCATCCGTACAATGGCGCGTGGGGTCCTATAAGTTGGGGCCACGCAGTCAGCGATAATCTTGTCGACTGGACGTTTTATCCTCCAGCCCTCATCCCCGGCGAGCTCTACGACAAACACGGATGCTTATCAGGATCCGGCATCGCCCACAACGGGTACCTGGTGCTGTTCTACACTGGTAATGCCGTATCAGACAACGCGACGCTCCAAACGCAAAACGTGGCGATCAGCACAGATGGTATCGTATTTCAGAAGTATATTTACAATCCTGTTATTCGGGACGGAGCTTTTGGGGCGGAGGACTCGCGAAACCCTAAAGTTTGGAGGTTCAGGAATGTCTGGTACATGCTCCTCACTAACACCCGGGAAGGGGTCGGCCGGCTTCTCCTCTACACGTCCATGGACCTTTTCAATTGGAAATTGAATGGCACTCTCGCACTGTCACTCGGCGACACCGGATATGCGTGGGAAAGTCCGGATTTGATCGAAATAGATGGTCAGCACGTTTTGATGTTGTGCCTCCAAGGGGTGCCTAGCGATGGATTTCGATTTAAAAATTTGTATCAAACTGGTTATATCGTCACTAATTTCAACTATGTTAATGGCCAGTTTGATGATTTAGAAGTCTCCACGGCAACATTTACGGAACTTGATCATGGACACGATTTTTATGCCCCGAAAACAGTGCTTGCTGTCGACGGCAGAAGACTCCTGATTGGCTGGCTCGGAATGTGGGAGAGCCATTTCAAAGAATCAAAGCACGGATGGGCCAGCATGCTCACCATTGTACGAGAAATGAAGCTGACCCCACAAGGTCGTCTCCTGATGCCTCCTGTTAGGGAAGTAGCTGAATTACGGACCGAAATACTTGAAGACGCGTGGTACAACCCCGGCGAGGCTTTCTATGCAGGGACGAGGACTTTTGAGTTGATAGTCAGCACTGCCAAAGTGTTTTACGATGTCGCCGTCATATTTGAATGGGATGGCGACCAGCAGTACACGGTGGGGTACTCGGCGGATCGAGGTCACATCATAGTGGATCGCGGCGGAGTGGACGGCCTCAGGAGGGCGGACTGGGCTCCAAACGATAAACTAAAACTGCGTATATTCGTCGACTATAGTTCAATAGAGGTTTTCTGTGGTTCCGGAGACGTGGTATTCTCCAGCCGCGTCTACCCAAAGAAAAATATCCGTGTGAAGATATCAGGAGAGTCACAGTTGCACGTCGTCCAGTATAAACTGCGACGCAGCGTCGGATATGATAGCAAATTGCGTAAATACTTGAAAGAGCATGTTCTCGAAAGGGCATAA
Protein
MDVLVVALYLRFVCAVFAVDLSHIETFVQENRYCLKQRYRPLYHISAAHAWTNHPSAFVYYKRQYHIFYQYHPYNGAWGPISWGHAVSDNLVDWTFYPPALIPGELYDKHGCLSGSGIAHNGYLVLFYTGNAVSDNATLQTQNVAISTDGIVFQKYIYNPVIRDGAFGAEDSRNPKVWRFRNVWYMLLTNTREGVGRLLLYTSMDLFNWKLNGTLALSLGDTGYAWESPDLIEIDGQHVLMLCLQGVPSDGFRFKNLYQTGYIVTNFNYVNGQFDDLEVSTATFTELDHGHDFYAPKTVLAVDGRRLLIGWLGMWESHFKESKHGWASMLTIVREMKLTPQGRLLMPPVREVAELRTEILEDAWYNPGEAFYAGTRTFELIVSTAKVFYDVAVIFEWDGDQQYTVGYSADRGHIIVDRGGVDGLRRADWAPNDKLKLRIFVDYSSIEVFCGSGDVVFSSRVYPKKNIRVKISGESQLHVVQYKLRRSVGYDSKLRKYLKEHVLERA
Summary
Description
Enables the bacterium to metabolize sucrose as a sole carbon source.
Catalytic Activity
Hydrolysis of terminal non-reducing beta-D-fructofuranoside residues in beta-D-fructofuranosides.
Similarity
Belongs to the glycosyl hydrolase 32 family.
Feature
chain Sucrose-6-phosphate hydrolase
Uniprot
B2DD58
A0A2H1WCL6
A0A2A4JK37
A0A194QXM3
A0A194QGN4
C9S280
+ More
L7X1F0 A0A0L7L2L9 A0A212EIA1 D1KRK8 A0A2M9NXW0 A0A2A4K783 A0A268DSW4 B2DD57 A0A264YVA8 A0A0M1NH34 A0A0L7L5N0 A0A2V1Z1H0 A0A369C7U8 A0A2H1WS64 W7ZAQ9 A0A2A8FJR6 A0A3G8Z3V4 S6FPW6 I2HVM8 A0A2K9K6V6 A0A0X8KHR7 A0A1N7FFJ1 A0A1D9PQ46 A0A2S9WLS3 A0A109N2J2 A0A172ZG79 A0A2H3M5E8 A0A270AEQ6 A0A3A5I7A1 A0A2M8SZV4 A0A0U0CY11 A0A2S4EM02 I2CAH2 A0A235BGJ0 A0A1U3VBD7 A7Z936 A0A0A0TW88 A0A1Y0XCW8 A0A0D7XT41 A0A1W6HM34
L7X1F0 A0A0L7L2L9 A0A212EIA1 D1KRK8 A0A2M9NXW0 A0A2A4K783 A0A268DSW4 B2DD57 A0A264YVA8 A0A0M1NH34 A0A0L7L5N0 A0A2V1Z1H0 A0A369C7U8 A0A2H1WS64 W7ZAQ9 A0A2A8FJR6 A0A3G8Z3V4 S6FPW6 I2HVM8 A0A2K9K6V6 A0A0X8KHR7 A0A1N7FFJ1 A0A1D9PQ46 A0A2S9WLS3 A0A109N2J2 A0A172ZG79 A0A2H3M5E8 A0A270AEQ6 A0A3A5I7A1 A0A2M8SZV4 A0A0U0CY11 A0A2S4EM02 I2CAH2 A0A235BGJ0 A0A1U3VBD7 A7Z936 A0A0A0TW88 A0A1Y0XCW8 A0A0D7XT41 A0A1W6HM34
EC Number
3.2.1.26
Pubmed
EMBL
BABH01020267
AB366560
BAG30883.1
ODYU01007762
SOQ50810.1
NWSH01001193
+ More
PCG72189.1 KQ461155 KPJ08326.1 KQ459232 KPJ02606.1 CU367882 CBH09256.1 KC469893 AGC92681.1 JTDY01003325 KOB69738.1 AGBW02014684 OWR41190.1 GQ293364 ACX49763.1 PHNG01000688 PJN90135.1 NWSH01000095 PCG79623.1 NPCI01000015 PAD63965.1 BABH01039865 AB366559 BAG30882.1 NOXE01000008 OZT12043.1 LIUS01000003 KOR81486.1 JTDY01002835 KOB70626.1 QGGK01000004 PWK00072.1 QPJZ01000008 RCX30090.1 ODYU01010661 SOQ55915.1 BAWA01000002 GAF11384.1 NUBA01000029 PEO46488.1 HG328253 CDG31284.1 AJST01000001 EIF14787.1 CP025001 AUJ75901.1 CP013950 AME07320.1 FTNS01000002 SIR99082.1 PVXB01000003 PRP64437.1 LNNH01000004 KWW22325.1 CP013023 ANF96277.1 NVNR01000012 PEF40132.1 NCWW01000025 PAL10818.1 NQYD01000001 RJS56667.1 PHIE01000003 PJH93070.1 CQZO01000003 COC83393.1 PQCN01000004 POO69139.1 CP003332 AFJ63646.1 CP018133 PTTN01000088 CP034203 ATU28215.1 PQB09528.1 QAR58347.1 FVSM01000016 SLC33593.1 CP000560 ABS75512.1 CP009748 AIW35302.1 CP021505 ARW40660.1 JZDI01000003 KJD58756.1 CP020893 ARM29293.1
PCG72189.1 KQ461155 KPJ08326.1 KQ459232 KPJ02606.1 CU367882 CBH09256.1 KC469893 AGC92681.1 JTDY01003325 KOB69738.1 AGBW02014684 OWR41190.1 GQ293364 ACX49763.1 PHNG01000688 PJN90135.1 NWSH01000095 PCG79623.1 NPCI01000015 PAD63965.1 BABH01039865 AB366559 BAG30882.1 NOXE01000008 OZT12043.1 LIUS01000003 KOR81486.1 JTDY01002835 KOB70626.1 QGGK01000004 PWK00072.1 QPJZ01000008 RCX30090.1 ODYU01010661 SOQ55915.1 BAWA01000002 GAF11384.1 NUBA01000029 PEO46488.1 HG328253 CDG31284.1 AJST01000001 EIF14787.1 CP025001 AUJ75901.1 CP013950 AME07320.1 FTNS01000002 SIR99082.1 PVXB01000003 PRP64437.1 LNNH01000004 KWW22325.1 CP013023 ANF96277.1 NVNR01000012 PEF40132.1 NCWW01000025 PAL10818.1 NQYD01000001 RJS56667.1 PHIE01000003 PJH93070.1 CQZO01000003 COC83393.1 PQCN01000004 POO69139.1 CP003332 AFJ63646.1 CP018133 PTTN01000088 CP034203 ATU28215.1 PQB09528.1 QAR58347.1 FVSM01000016 SLC33593.1 CP000560 ABS75512.1 CP009748 AIW35302.1 CP021505 ARW40660.1 JZDI01000003 KJD58756.1 CP020893 ARM29293.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000231794 UP000215585 UP000242943 UP000036875 UP000245756 UP000253302 UP000019372 UP000219857 UP000015481 UP000005799 UP000234366 UP000061465 UP000186598 UP000238133 UP000064189 UP000078148 UP000220819 UP000215626 UP000279300 UP000229353 UP000039962 UP000002878 UP000228760 UP000239383 UP000287786 UP000249266 UP000001120 UP000030075 UP000195486 UP000032472 UP000193819
UP000231794 UP000215585 UP000242943 UP000036875 UP000245756 UP000253302 UP000019372 UP000219857 UP000015481 UP000005799 UP000234366 UP000061465 UP000186598 UP000238133 UP000064189 UP000078148 UP000220819 UP000215626 UP000279300 UP000229353 UP000039962 UP000002878 UP000228760 UP000239383 UP000287786 UP000249266 UP000001120 UP000030075 UP000195486 UP000032472 UP000193819
Interpro
Gene 3D
ProteinModelPortal
B2DD58
A0A2H1WCL6
A0A2A4JK37
A0A194QXM3
A0A194QGN4
C9S280
+ More
L7X1F0 A0A0L7L2L9 A0A212EIA1 D1KRK8 A0A2M9NXW0 A0A2A4K783 A0A268DSW4 B2DD57 A0A264YVA8 A0A0M1NH34 A0A0L7L5N0 A0A2V1Z1H0 A0A369C7U8 A0A2H1WS64 W7ZAQ9 A0A2A8FJR6 A0A3G8Z3V4 S6FPW6 I2HVM8 A0A2K9K6V6 A0A0X8KHR7 A0A1N7FFJ1 A0A1D9PQ46 A0A2S9WLS3 A0A109N2J2 A0A172ZG79 A0A2H3M5E8 A0A270AEQ6 A0A3A5I7A1 A0A2M8SZV4 A0A0U0CY11 A0A2S4EM02 I2CAH2 A0A235BGJ0 A0A1U3VBD7 A7Z936 A0A0A0TW88 A0A1Y0XCW8 A0A0D7XT41 A0A1W6HM34
L7X1F0 A0A0L7L2L9 A0A212EIA1 D1KRK8 A0A2M9NXW0 A0A2A4K783 A0A268DSW4 B2DD57 A0A264YVA8 A0A0M1NH34 A0A0L7L5N0 A0A2V1Z1H0 A0A369C7U8 A0A2H1WS64 W7ZAQ9 A0A2A8FJR6 A0A3G8Z3V4 S6FPW6 I2HVM8 A0A2K9K6V6 A0A0X8KHR7 A0A1N7FFJ1 A0A1D9PQ46 A0A2S9WLS3 A0A109N2J2 A0A172ZG79 A0A2H3M5E8 A0A270AEQ6 A0A3A5I7A1 A0A2M8SZV4 A0A0U0CY11 A0A2S4EM02 I2CAH2 A0A235BGJ0 A0A1U3VBD7 A7Z936 A0A0A0TW88 A0A1Y0XCW8 A0A0D7XT41 A0A1W6HM34
PDB
1UYP
E-value=1.18603e-49,
Score=497
Ontologies
GO
Topology
Subcellular location
Cytoplasm
SignalP
Position: 1 - 18,
Likelihood: 0.743938
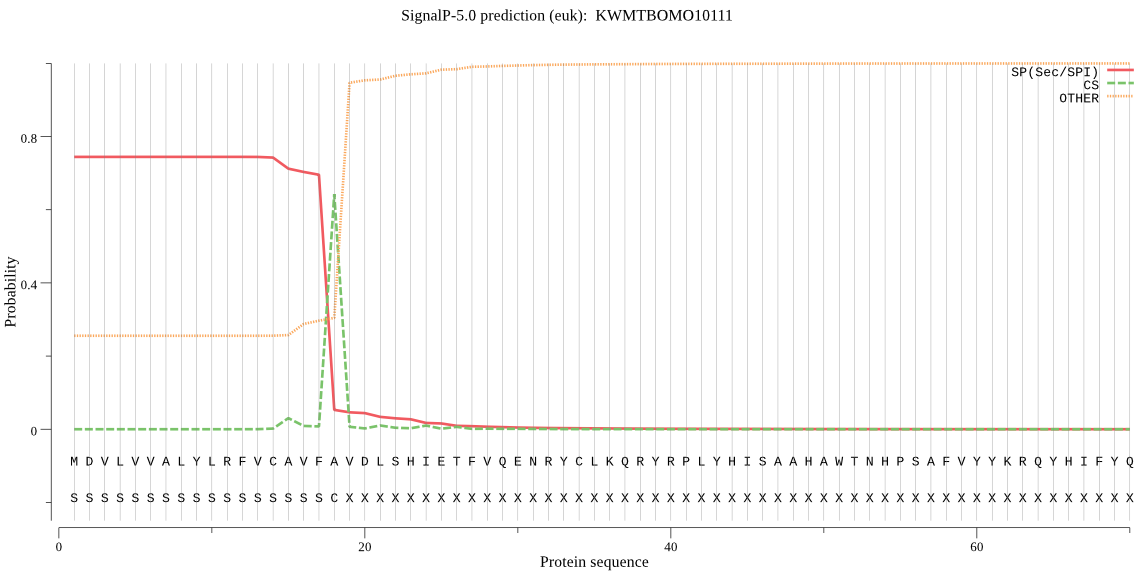
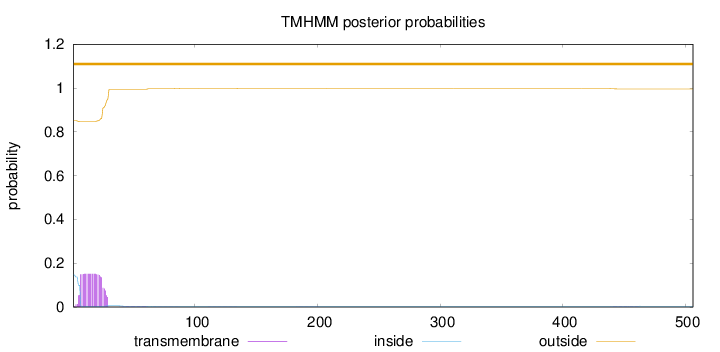
Length:
506
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.28958000000001
Exp number, first 60 AAs:
3.22764
Total prob of N-in:
0.14862
outside
1 - 506
Population Genetic Test Statistics
Pi
258.912024
Theta
199.793761
Tajima's D
0.993406
CLR
0.017084
CSRT
0.661816909154542
Interpretation
Uncertain
