Gene
KWMTBOMO10110
Pre Gene Modal
BGIBMGA005553
Annotation
PREDICTED:_sorting_nexin-8-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.982 Nuclear Reliability : 1.381
Sequence
CDS
ATGACAAATACAAATGAAGTAACATTTCAAGAGCTTGAAGCAACTGATGTTGTAAGCATAGACCTTGTACCCGAACGGAAGGGAATTATACTAAAACACTGCGAGTATTATGTCAGCTCAAGGCGTCATGGCACAACTGTGACCAGAAGATACAATGATTTTGTACAGCTTTACGATGTGCTATTTGCTAAATATCCTTATCGGGCTGTATGTCAGCTACCTCCGAAACGTGTGGTGGTTGGGGGCGGAAGTGGACAGTTCTTAGAATCCCGTCGCGCTGCTCTCCAACGTTGGCTATCGTTGGCCGCCTGTCATCCAATATTGGCGCATGATGCTGACTTGCGCATCTTCCTATCTGAGTCGGTCCTGCGGCTGGAGAAACCTAAGCACGACGAGTTCGTGCTTGCCGGTACTCAGTGCGATGAACAGGAAGAAACATCTCTAGACGAAATGAAGGCGAGGTTTGCGAAGGAGCAGGAACAACTGCGGATCTTGTACTTGGGGATGGATAGACTAGCGCAGATATTCGATAGAGTTAAAATCAGATGCGAAGCAGAAAAATCAGATTTCCGTGAGCTGGGGGCCGCGCTGCAGGCGCTGTCGACGCGGGACGCCGCCGACACCCCGCTCTGGACCCGGCTCAAGCAAGCTTACGGCGACGCGGCGCTGGTATGTACCGGGGAGATGTTCGACGACGAGTCTGTGGACAGCGAGGCGGAGGGGCGGGTGTCGCTGGCGCGCGACGCGCTGGCGGCCGGCCGCGAGCTGTGCGCGCGGCTGGGGCGCGGCCTGCACGCAGAGCGCGCCGCCGCCGCCGCGCTCGCGCCGCCCCACGCGCAGCTGCGGAGACGACACCTCTACGCGCTCACCGCCGTCCTTCAGTTTTTCCAAATCGATGATGAATTACAAAATTTTCATCAAACTAAACCATAA
Protein
MTNTNEVTFQELEATDVVSIDLVPERKGIILKHCEYYVSSRRHGTTVTRRYNDFVQLYDVLFAKYPYRAVCQLPPKRVVVGGGSGQFLESRRAALQRWLSLAACHPILAHDADLRIFLSESVLRLEKPKHDEFVLAGTQCDEQEETSLDEMKARFAKEQEQLRILYLGMDRLAQIFDRVKIRCEAEKSDFRELGAALQALSTRDAADTPLWTRLKQAYGDAALVCTGEMFDDESVDSEAEGRVSLARDALAAGRELCARLGRGLHAERAAAAALAPPHAQLRRRHLYALTAVLQFFQIDDELQNFHQTKP
Summary
Uniprot
C9S279
A0A194QC95
A0A212FII3
L7X3V4
A0A0L7L2U4
A0A0V0G7R4
+ More
T1IAK3 A0A067QJI4 A0A1B6K236 A0A2J7QEC3 A0A1B6MBK3 A0A0P4VG87 A0A0A9XXK9 A0A1B6EQ61 A0A026WXX4 E9JBZ6 A0A2R7WT69 A0A158NKL6 A0A195F594 A0A195D027 E2AYD4 A0A195EJ76 A0A0J7KP47 A0A195BVX5 A0A310SCI9 A0A2A3E843 V9IEV8 A0A088AEF3 F4WD10 A0A3L8E1W0 E2BQQ8 A0A0L7QUM0 A0A0N0BCG6 A0A2B4S240 A0A091ER28 A0A0B6XYZ4 A0A1W7RFE0 A0A3M0KWP6 A0A091GHJ3 A0A0F7Z5K8 A0A099ZXJ9 A0A091GZW2 H0ZCW4 T1E5N7 A0A099ZG95 K7FRX3 U3KHL0 A0A3L8SFM6 H9GEQ0 A0A218V7J9 A0A091V9R0 A0A091JPU5 A0A091T427 A0A091QGY1 A0A091JVH7 A0A093SP84 A0A3Q2U9X6 G1MUT0 A0A091IAN8 A0A0Q3XBA6 A0A093J7S8 A0A091V764 A0A091WAY1 A0A091TX84 A0A091KXT8 A0A091MHX5 R0JUU4 A0A093DQ12 U3J8J4 A0A2R5LKU5 A0A093J9L5 A0A093NW47 A0A087RFX3 A0A2I0M0P8 A0A3Q2U5T3 A0A091NKS3 A0A1W4X8J3 V4C0G1 A0A093R2D6 F1NZ35 A0A1V4KSA8 A0A293L3X6 E2R8Q0 A0A293MEI5
T1IAK3 A0A067QJI4 A0A1B6K236 A0A2J7QEC3 A0A1B6MBK3 A0A0P4VG87 A0A0A9XXK9 A0A1B6EQ61 A0A026WXX4 E9JBZ6 A0A2R7WT69 A0A158NKL6 A0A195F594 A0A195D027 E2AYD4 A0A195EJ76 A0A0J7KP47 A0A195BVX5 A0A310SCI9 A0A2A3E843 V9IEV8 A0A088AEF3 F4WD10 A0A3L8E1W0 E2BQQ8 A0A0L7QUM0 A0A0N0BCG6 A0A2B4S240 A0A091ER28 A0A0B6XYZ4 A0A1W7RFE0 A0A3M0KWP6 A0A091GHJ3 A0A0F7Z5K8 A0A099ZXJ9 A0A091GZW2 H0ZCW4 T1E5N7 A0A099ZG95 K7FRX3 U3KHL0 A0A3L8SFM6 H9GEQ0 A0A218V7J9 A0A091V9R0 A0A091JPU5 A0A091T427 A0A091QGY1 A0A091JVH7 A0A093SP84 A0A3Q2U9X6 G1MUT0 A0A091IAN8 A0A0Q3XBA6 A0A093J7S8 A0A091V764 A0A091WAY1 A0A091TX84 A0A091KXT8 A0A091MHX5 R0JUU4 A0A093DQ12 U3J8J4 A0A2R5LKU5 A0A093J9L5 A0A093NW47 A0A087RFX3 A0A2I0M0P8 A0A3Q2U5T3 A0A091NKS3 A0A1W4X8J3 V4C0G1 A0A093R2D6 F1NZ35 A0A1V4KSA8 A0A293L3X6 E2R8Q0 A0A293MEI5
Pubmed
EMBL
CU367882
CBH09255.1
KQ459232
KPJ02605.1
AGBW02008375
OWR53549.1
+ More
KC469893 AGC92680.2 JTDY01003325 KOB69740.1 GECL01002107 JAP04017.1 ACPB03000155 KK853286 KDR08951.1 GECU01002177 JAT05530.1 NEVH01015316 PNF26938.1 GEBQ01006680 JAT33297.1 GDKW01003409 JAI53186.1 GBHO01018062 GBRD01013395 GDHC01009049 JAG25542.1 JAG52431.1 JAQ09580.1 GECZ01029816 JAS39953.1 KK107087 EZA59994.1 GL771642 EFZ09670.1 KK855480 PTY22756.1 ADTU01002109 ADTU01002110 ADTU01002111 KQ981805 KYN35566.1 KQ977110 KYN05729.1 GL443887 EFN61551.1 KQ978801 KYN28315.1 LBMM01004735 KMQ92123.1 KQ976403 KYM92116.1 KQ770367 OAD52656.1 KZ288355 PBC27221.1 JR040589 AEY59197.1 GL888077 EGI67917.1 QOIP01000001 RLU26443.1 GL449798 EFN81977.1 KQ414734 KOC62337.1 KQ435908 KOX68968.1 LSMT01000234 PFX22628.1 KK718885 KFO59037.1 HACG01002213 CEK49078.1 GDAY02001586 JAV49841.1 QRBI01000098 RMC17658.1 KL448274 KFO81810.1 GBEX01002225 JAI12335.1 KL869946 KGL86571.1 KL516011 KFO88062.1 ABQF01038220 GAAZ01001611 GBKC01002367 JAA96332.1 JAG43703.1 KL892473 KGL79845.1 AGCU01017993 AGCU01017994 AGCU01017995 AGCU01017996 AGTO01010793 QUSF01000022 RLW01412.1 AAWZ02025450 MUZQ01000034 OWK61916.1 KL410807 KFQ99848.1 KK502235 KFP21770.1 KK496095 KFQ65656.1 KK698712 KFQ25962.1 KK531214 KFP28091.1 KL672051 KFW84434.1 AADN05000376 KL218366 KFP04538.1 LMAW01000001 KQL61485.1 KK607530 KFW10008.1 KK417028 KFQ85563.1 KK735067 KFR12265.1 KK468089 KFQ83044.1 KK758897 KFP44248.1 KK828391 KFP74543.1 KB743145 EOB00952.1 KL228800 KFV04251.1 ADON01076621 GGLE01006024 MBY10150.1 KK578004 KFW11720.1 KL224810 KFW64357.1 KL226347 KFM12377.1 AKCR02000052 PKK23254.1 KL387376 KFP90373.1 KB201721 ESO94914.1 KL426122 KFW90342.1 LSYS01001700 OPJ87313.1 GFWV01000397 MAA25127.1 AAEX03004329 GFWV01013850 MAA38579.1
KC469893 AGC92680.2 JTDY01003325 KOB69740.1 GECL01002107 JAP04017.1 ACPB03000155 KK853286 KDR08951.1 GECU01002177 JAT05530.1 NEVH01015316 PNF26938.1 GEBQ01006680 JAT33297.1 GDKW01003409 JAI53186.1 GBHO01018062 GBRD01013395 GDHC01009049 JAG25542.1 JAG52431.1 JAQ09580.1 GECZ01029816 JAS39953.1 KK107087 EZA59994.1 GL771642 EFZ09670.1 KK855480 PTY22756.1 ADTU01002109 ADTU01002110 ADTU01002111 KQ981805 KYN35566.1 KQ977110 KYN05729.1 GL443887 EFN61551.1 KQ978801 KYN28315.1 LBMM01004735 KMQ92123.1 KQ976403 KYM92116.1 KQ770367 OAD52656.1 KZ288355 PBC27221.1 JR040589 AEY59197.1 GL888077 EGI67917.1 QOIP01000001 RLU26443.1 GL449798 EFN81977.1 KQ414734 KOC62337.1 KQ435908 KOX68968.1 LSMT01000234 PFX22628.1 KK718885 KFO59037.1 HACG01002213 CEK49078.1 GDAY02001586 JAV49841.1 QRBI01000098 RMC17658.1 KL448274 KFO81810.1 GBEX01002225 JAI12335.1 KL869946 KGL86571.1 KL516011 KFO88062.1 ABQF01038220 GAAZ01001611 GBKC01002367 JAA96332.1 JAG43703.1 KL892473 KGL79845.1 AGCU01017993 AGCU01017994 AGCU01017995 AGCU01017996 AGTO01010793 QUSF01000022 RLW01412.1 AAWZ02025450 MUZQ01000034 OWK61916.1 KL410807 KFQ99848.1 KK502235 KFP21770.1 KK496095 KFQ65656.1 KK698712 KFQ25962.1 KK531214 KFP28091.1 KL672051 KFW84434.1 AADN05000376 KL218366 KFP04538.1 LMAW01000001 KQL61485.1 KK607530 KFW10008.1 KK417028 KFQ85563.1 KK735067 KFR12265.1 KK468089 KFQ83044.1 KK758897 KFP44248.1 KK828391 KFP74543.1 KB743145 EOB00952.1 KL228800 KFV04251.1 ADON01076621 GGLE01006024 MBY10150.1 KK578004 KFW11720.1 KL224810 KFW64357.1 KL226347 KFM12377.1 AKCR02000052 PKK23254.1 KL387376 KFP90373.1 KB201721 ESO94914.1 KL426122 KFW90342.1 LSYS01001700 OPJ87313.1 GFWV01000397 MAA25127.1 AAEX03004329 GFWV01013850 MAA38579.1
Proteomes
UP000053268
UP000007151
UP000037510
UP000015103
UP000027135
UP000235965
+ More
UP000053097 UP000005205 UP000078541 UP000078542 UP000000311 UP000078492 UP000036403 UP000078540 UP000242457 UP000005203 UP000007755 UP000279307 UP000008237 UP000053825 UP000053105 UP000225706 UP000052976 UP000269221 UP000053760 UP000053858 UP000007754 UP000053641 UP000007267 UP000016665 UP000276834 UP000001646 UP000197619 UP000053283 UP000053119 UP000053258 UP000000539 UP000001645 UP000054308 UP000051836 UP000053605 UP000016666 UP000054081 UP000053286 UP000053872 UP000192223 UP000030746 UP000190648 UP000002254
UP000053097 UP000005205 UP000078541 UP000078542 UP000000311 UP000078492 UP000036403 UP000078540 UP000242457 UP000005203 UP000007755 UP000279307 UP000008237 UP000053825 UP000053105 UP000225706 UP000052976 UP000269221 UP000053760 UP000053858 UP000007754 UP000053641 UP000007267 UP000016665 UP000276834 UP000001646 UP000197619 UP000053283 UP000053119 UP000053258 UP000000539 UP000001645 UP000054308 UP000051836 UP000053605 UP000016666 UP000054081 UP000053286 UP000053872 UP000192223 UP000030746 UP000190648 UP000002254
PRIDE
Pfam
PF00787 PX
Interpro
Gene 3D
ProteinModelPortal
C9S279
A0A194QC95
A0A212FII3
L7X3V4
A0A0L7L2U4
A0A0V0G7R4
+ More
T1IAK3 A0A067QJI4 A0A1B6K236 A0A2J7QEC3 A0A1B6MBK3 A0A0P4VG87 A0A0A9XXK9 A0A1B6EQ61 A0A026WXX4 E9JBZ6 A0A2R7WT69 A0A158NKL6 A0A195F594 A0A195D027 E2AYD4 A0A195EJ76 A0A0J7KP47 A0A195BVX5 A0A310SCI9 A0A2A3E843 V9IEV8 A0A088AEF3 F4WD10 A0A3L8E1W0 E2BQQ8 A0A0L7QUM0 A0A0N0BCG6 A0A2B4S240 A0A091ER28 A0A0B6XYZ4 A0A1W7RFE0 A0A3M0KWP6 A0A091GHJ3 A0A0F7Z5K8 A0A099ZXJ9 A0A091GZW2 H0ZCW4 T1E5N7 A0A099ZG95 K7FRX3 U3KHL0 A0A3L8SFM6 H9GEQ0 A0A218V7J9 A0A091V9R0 A0A091JPU5 A0A091T427 A0A091QGY1 A0A091JVH7 A0A093SP84 A0A3Q2U9X6 G1MUT0 A0A091IAN8 A0A0Q3XBA6 A0A093J7S8 A0A091V764 A0A091WAY1 A0A091TX84 A0A091KXT8 A0A091MHX5 R0JUU4 A0A093DQ12 U3J8J4 A0A2R5LKU5 A0A093J9L5 A0A093NW47 A0A087RFX3 A0A2I0M0P8 A0A3Q2U5T3 A0A091NKS3 A0A1W4X8J3 V4C0G1 A0A093R2D6 F1NZ35 A0A1V4KSA8 A0A293L3X6 E2R8Q0 A0A293MEI5
T1IAK3 A0A067QJI4 A0A1B6K236 A0A2J7QEC3 A0A1B6MBK3 A0A0P4VG87 A0A0A9XXK9 A0A1B6EQ61 A0A026WXX4 E9JBZ6 A0A2R7WT69 A0A158NKL6 A0A195F594 A0A195D027 E2AYD4 A0A195EJ76 A0A0J7KP47 A0A195BVX5 A0A310SCI9 A0A2A3E843 V9IEV8 A0A088AEF3 F4WD10 A0A3L8E1W0 E2BQQ8 A0A0L7QUM0 A0A0N0BCG6 A0A2B4S240 A0A091ER28 A0A0B6XYZ4 A0A1W7RFE0 A0A3M0KWP6 A0A091GHJ3 A0A0F7Z5K8 A0A099ZXJ9 A0A091GZW2 H0ZCW4 T1E5N7 A0A099ZG95 K7FRX3 U3KHL0 A0A3L8SFM6 H9GEQ0 A0A218V7J9 A0A091V9R0 A0A091JPU5 A0A091T427 A0A091QGY1 A0A091JVH7 A0A093SP84 A0A3Q2U9X6 G1MUT0 A0A091IAN8 A0A0Q3XBA6 A0A093J7S8 A0A091V764 A0A091WAY1 A0A091TX84 A0A091KXT8 A0A091MHX5 R0JUU4 A0A093DQ12 U3J8J4 A0A2R5LKU5 A0A093J9L5 A0A093NW47 A0A087RFX3 A0A2I0M0P8 A0A3Q2U5T3 A0A091NKS3 A0A1W4X8J3 V4C0G1 A0A093R2D6 F1NZ35 A0A1V4KSA8 A0A293L3X6 E2R8Q0 A0A293MEI5
PDB
6H7W
E-value=2.80002e-07,
Score=129
Ontologies
KEGG
GO
PANTHER
Topology
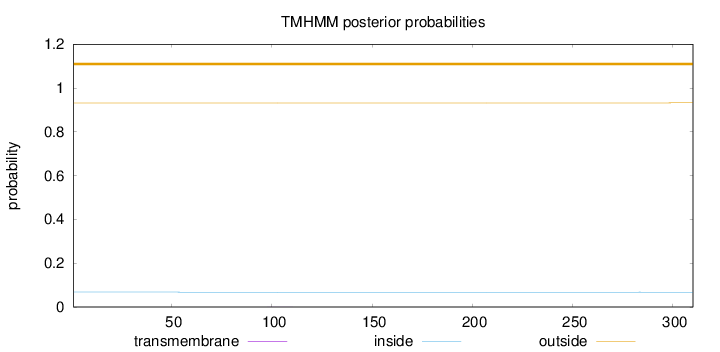
Length:
310
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00601999999999999
Exp number, first 60 AAs:
0.00052
Total prob of N-in:
0.06797
outside
1 - 310
Population Genetic Test Statistics
Pi
177.527904
Theta
153.783717
Tajima's D
0.066214
CLR
111.278713
CSRT
0.390930453477326
Interpretation
Uncertain
