Gene
KWMTBOMO10102
Pre Gene Modal
BGIBMGA005659
Annotation
PREDICTED:_uncharacterized_protein_LOC101746409_isoform_X2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.828
Sequence
CDS
ATGTTGAGAAATCTAAATTATTTAAGGCATTGTTTTCGTGTTAGCGGAATTAGATTTAAAAGCAGCAATGTTGAAATTAACGAAAATGAACCGGTAAAATTTTCAACGAGCAGAGCTGCCTCTAGAGGTCCCGTTCCTTTAATCAGCAAAATAAATGATGATATGCCTTGGTACCAACCGTACGTAGTCATCGGAAGCGTTGCAATCTTCATGTTATATTTTTGTGTTTTGAGGGAAGAAAGTGACATTGACCGGGAATTCGACAAAACACTTTATGATAGGATAAAAGGCTTAGAAAAAGAACAACTCCTCCAATCATACAGATACAACAAAGAACATGGTAAAAGTGTTATTGAAATCGAAGAGAGATTGCTGGAAATTGAGAAGGCAGAGAAGGAACTACAGCTTTGA
Protein
MLRNLNYLRHCFRVSGIRFKSSNVEINENEPVKFSTSRAASRGPVPLISKINDDMPWYQPYVVIGSVAIFMLYFCVLREESDIDREFDKTLYDRIKGLEKEQLLQSYRYNKEHGKSVIEIEERLLEIEKAEKELQL
Summary
Uniprot
H9J817
A0A194QXM8
A0A2H1X5Q7
A0A194QGM9
A0A0L7L3N1
A0A2A4JJB1
+ More
L7X1E5 S4PWX3 B0WVH4 B4MRE3 B4HRL0 A0A0J9TW40 B4QFP3 B4NS83 A0A1L8EBT8 A0A1L8ECD6 A0A1L8EBU5 A0A1L8EBT0 A1Z7B8 A0A1B0AP75 Q8IGB5 A0A1W4WB57 A0A0K8UDX3 A0A1S4FPF7 Q16TY0 A0A023EGN2 A0A182H8Q1 A0A0K8V407 A0A034WJ88 A0A0C9RIS6 J3JXE1 A0A0M4EU57 B4J5L9 A0A0P4WC84 B4P2P4 A0A182WM28 A0A182REM1 A0A2M3Z994 A0A2M3Z971 A0A182JJ20 A0A182T598 A0A182MP43 A0A182YQD3 A0A084WDZ9 A0A182V1F5 A0A182XJJ3 A0A182HQM2 Q7Q7X9 A0A182JPH4 A0NB56 A0A182PGB6 A0A182NW70 A0A182TQ52 A0A2M4AYG3 A0A0P6BUF5 V5GRF9 A0A1L8D923 A0A1L8D8S6 A0A2J7PKN7 A0A1D2NFM9 A0A2T7Q1K6 A0A0U2V658
L7X1E5 S4PWX3 B0WVH4 B4MRE3 B4HRL0 A0A0J9TW40 B4QFP3 B4NS83 A0A1L8EBT8 A0A1L8ECD6 A0A1L8EBU5 A0A1L8EBT0 A1Z7B8 A0A1B0AP75 Q8IGB5 A0A1W4WB57 A0A0K8UDX3 A0A1S4FPF7 Q16TY0 A0A023EGN2 A0A182H8Q1 A0A0K8V407 A0A034WJ88 A0A0C9RIS6 J3JXE1 A0A0M4EU57 B4J5L9 A0A0P4WC84 B4P2P4 A0A182WM28 A0A182REM1 A0A2M3Z994 A0A2M3Z971 A0A182JJ20 A0A182T598 A0A182MP43 A0A182YQD3 A0A084WDZ9 A0A182V1F5 A0A182XJJ3 A0A182HQM2 Q7Q7X9 A0A182JPH4 A0NB56 A0A182PGB6 A0A182NW70 A0A182TQ52 A0A2M4AYG3 A0A0P6BUF5 V5GRF9 A0A1L8D923 A0A1L8D8S6 A0A2J7PKN7 A0A1D2NFM9 A0A2T7Q1K6 A0A0U2V658
Pubmed
EMBL
BABH01020273
KQ461155
KPJ08331.1
ODYU01013238
SOQ59934.1
KQ459232
+ More
KPJ02601.1 JTDY01003237 KOB69899.1 NWSH01001193 PCG72185.1 KC469893 AGC92676.1 GAIX01006038 JAA86522.1 DS232126 EDS35580.1 CH963850 EDW74682.1 CH480816 EDW46892.1 CM002911 KMY92220.1 CM000362 EDX06149.1 CH981979 EDX15461.1 GFDG01002589 JAV16210.1 GFDG01002596 JAV16203.1 GFDG01002591 JAV16208.1 GFDG01002592 JAV16207.1 AE013599 KX531814 AAM68855.1 ANY27624.1 JXJN01001234 BT001862 AAN71628.1 GDHF01027442 JAI24872.1 CH477637 EAT37986.1 GAPW01005623 JAC07975.1 JXUM01119077 KQ566252 KXJ70279.1 GDHF01018718 GDHF01013343 GDHF01003597 JAI33596.1 JAI38971.1 JAI48717.1 GAKP01004530 JAC54422.1 GBYB01006791 JAG76558.1 APGK01035707 BT127910 KB740928 KB632428 AEE62872.1 ENN77921.1 ERL95410.1 CP012524 ALC40895.1 CH916367 EDW00782.1 GDRN01053048 JAI66205.1 CM000157 EDW89305.1 GGFM01004309 MBW25060.1 GGFM01004310 MBW25061.1 AXCM01002148 ATLV01023096 KE525340 KFB48443.1 APCN01004661 AAAB01008948 EAA10457.2 AAAB01008742 EAU77780.2 GGFK01012512 MBW45833.1 GDIP01017288 JAM86427.1 GALX01005618 JAB62848.1 GFDF01011237 JAV02847.1 GFDF01011236 JAV02848.1 NEVH01024535 PNF16895.1 LJIJ01000060 ODN03895.1 PZQS01000001 PVD39555.1 KT754667 ALS04501.1
KPJ02601.1 JTDY01003237 KOB69899.1 NWSH01001193 PCG72185.1 KC469893 AGC92676.1 GAIX01006038 JAA86522.1 DS232126 EDS35580.1 CH963850 EDW74682.1 CH480816 EDW46892.1 CM002911 KMY92220.1 CM000362 EDX06149.1 CH981979 EDX15461.1 GFDG01002589 JAV16210.1 GFDG01002596 JAV16203.1 GFDG01002591 JAV16208.1 GFDG01002592 JAV16207.1 AE013599 KX531814 AAM68855.1 ANY27624.1 JXJN01001234 BT001862 AAN71628.1 GDHF01027442 JAI24872.1 CH477637 EAT37986.1 GAPW01005623 JAC07975.1 JXUM01119077 KQ566252 KXJ70279.1 GDHF01018718 GDHF01013343 GDHF01003597 JAI33596.1 JAI38971.1 JAI48717.1 GAKP01004530 JAC54422.1 GBYB01006791 JAG76558.1 APGK01035707 BT127910 KB740928 KB632428 AEE62872.1 ENN77921.1 ERL95410.1 CP012524 ALC40895.1 CH916367 EDW00782.1 GDRN01053048 JAI66205.1 CM000157 EDW89305.1 GGFM01004309 MBW25060.1 GGFM01004310 MBW25061.1 AXCM01002148 ATLV01023096 KE525340 KFB48443.1 APCN01004661 AAAB01008948 EAA10457.2 AAAB01008742 EAU77780.2 GGFK01012512 MBW45833.1 GDIP01017288 JAM86427.1 GALX01005618 JAB62848.1 GFDF01011237 JAV02847.1 GFDF01011236 JAV02848.1 NEVH01024535 PNF16895.1 LJIJ01000060 ODN03895.1 PZQS01000001 PVD39555.1 KT754667 ALS04501.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000218220
UP000002320
+ More
UP000007798 UP000001292 UP000000304 UP000000803 UP000092460 UP000192221 UP000008820 UP000069940 UP000249989 UP000019118 UP000030742 UP000092553 UP000001070 UP000002282 UP000075920 UP000075900 UP000075880 UP000075901 UP000075883 UP000076408 UP000030765 UP000075903 UP000076407 UP000075840 UP000007062 UP000075881 UP000075885 UP000075884 UP000075902 UP000235965 UP000094527 UP000245119
UP000007798 UP000001292 UP000000304 UP000000803 UP000092460 UP000192221 UP000008820 UP000069940 UP000249989 UP000019118 UP000030742 UP000092553 UP000001070 UP000002282 UP000075920 UP000075900 UP000075880 UP000075901 UP000075883 UP000076408 UP000030765 UP000075903 UP000076407 UP000075840 UP000007062 UP000075881 UP000075885 UP000075884 UP000075902 UP000235965 UP000094527 UP000245119
Pfam
PF15013 CCSMST1
Interpro
IPR029160
CCSMST1_fam
ProteinModelPortal
H9J817
A0A194QXM8
A0A2H1X5Q7
A0A194QGM9
A0A0L7L3N1
A0A2A4JJB1
+ More
L7X1E5 S4PWX3 B0WVH4 B4MRE3 B4HRL0 A0A0J9TW40 B4QFP3 B4NS83 A0A1L8EBT8 A0A1L8ECD6 A0A1L8EBU5 A0A1L8EBT0 A1Z7B8 A0A1B0AP75 Q8IGB5 A0A1W4WB57 A0A0K8UDX3 A0A1S4FPF7 Q16TY0 A0A023EGN2 A0A182H8Q1 A0A0K8V407 A0A034WJ88 A0A0C9RIS6 J3JXE1 A0A0M4EU57 B4J5L9 A0A0P4WC84 B4P2P4 A0A182WM28 A0A182REM1 A0A2M3Z994 A0A2M3Z971 A0A182JJ20 A0A182T598 A0A182MP43 A0A182YQD3 A0A084WDZ9 A0A182V1F5 A0A182XJJ3 A0A182HQM2 Q7Q7X9 A0A182JPH4 A0NB56 A0A182PGB6 A0A182NW70 A0A182TQ52 A0A2M4AYG3 A0A0P6BUF5 V5GRF9 A0A1L8D923 A0A1L8D8S6 A0A2J7PKN7 A0A1D2NFM9 A0A2T7Q1K6 A0A0U2V658
L7X1E5 S4PWX3 B0WVH4 B4MRE3 B4HRL0 A0A0J9TW40 B4QFP3 B4NS83 A0A1L8EBT8 A0A1L8ECD6 A0A1L8EBU5 A0A1L8EBT0 A1Z7B8 A0A1B0AP75 Q8IGB5 A0A1W4WB57 A0A0K8UDX3 A0A1S4FPF7 Q16TY0 A0A023EGN2 A0A182H8Q1 A0A0K8V407 A0A034WJ88 A0A0C9RIS6 J3JXE1 A0A0M4EU57 B4J5L9 A0A0P4WC84 B4P2P4 A0A182WM28 A0A182REM1 A0A2M3Z994 A0A2M3Z971 A0A182JJ20 A0A182T598 A0A182MP43 A0A182YQD3 A0A084WDZ9 A0A182V1F5 A0A182XJJ3 A0A182HQM2 Q7Q7X9 A0A182JPH4 A0NB56 A0A182PGB6 A0A182NW70 A0A182TQ52 A0A2M4AYG3 A0A0P6BUF5 V5GRF9 A0A1L8D923 A0A1L8D8S6 A0A2J7PKN7 A0A1D2NFM9 A0A2T7Q1K6 A0A0U2V658
Ontologies
KEGG
GO
PANTHER
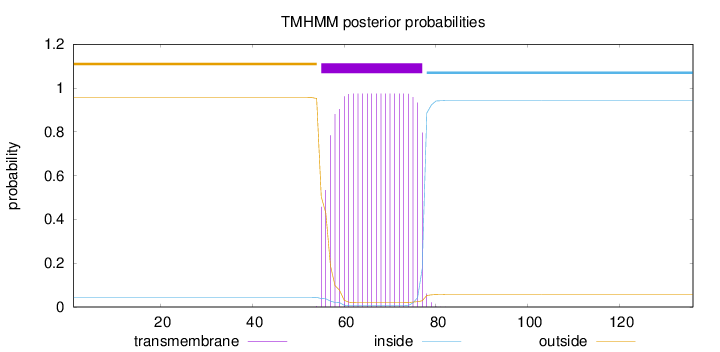
Topology
Length:
136
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.94573
Exp number, first 60 AAs:
4.52716
Total prob of N-in:
0.04376
outside
1 - 54
TMhelix
55 - 77
inside
78 - 136
Population Genetic Test Statistics
Pi
162.711973
Theta
154.00512
Tajima's D
0.130057
CLR
0.302821
CSRT
0.407929603519824
Interpretation
Uncertain
