Gene
KWMTBOMO10101
Pre Gene Modal
BGIBMGA005660
Annotation
RING_finger_protein_unkempt_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 2.263 Nuclear Reliability : 1.959
Sequence
CDS
ATGCCGTCGGAATCCAAGCCTTTGTTAACAGCCCAAACCGAAAAACCGAATCATTACAAATATCTAAAAGAATTCCGAGTGGAGCAGTGTCCATCTTTTCTCCAACATAAATGTACACAACACAGGCCGTTTACGTGTTTTCATTGGCATTTTAATAATCAAAGGAGACGCAGACCTGTTAGAAAAAGAGACGGGACCTTCAACTATAGCGCGGACAATTACTGTACGAAGTACAACGAGACGTCGGGCGTTTGCGAAGACGGAGACGAGTGTCCGTTCCTGCATCGTACAGCAGGCGACACGGAGCGACGATATCACTTGCGCTACTACAAAACTTGCATGTGCGTACACGACACGGACACGCGCGGCCTCTGCACCAAGAACGGGGCGCATTGCGCGTTCGCACACGGAGCGCCCGACCTGCGCCCCCCCGTGCTGGACGTGCGCGAGCTGCACGCACTCGACGCGGACGACGACGCCGCGCCCAACGCCCTCGACCGAGAGCGGAACCTCATCAACGAAGACCCCAAGTGGCAAGATACAAATTATGTCCTGTCATCATATAAAACGGAACCATGTAAAAGACCTCCTAGGCTTTGTAGACAAGGGTATGCATGCCCTCAGTATCATAATAGTAAAGATAAACGTAGGTCTCCTAGAAAGTATAAATATCGCAGCACACCATGCCCGAACGTGAAGCATGGTGAGGAGTGGGGCGAGCCCAGCAATTGTGAAGCCGGTGACACTTGTGGATACTGTCATACGCGGACGGAGCAGCAGTTCCATCCAGAAATCTATAAATCCACAAAATGCAACGACGTTCAGCAGGCGGGATACTGTCCACGAGGCTTGTTCTGCGCCTTTGCGCACGTTGAGCCCGAAGAACCTGGCGGCCGCGAGCCCGGCCCCGGCGACTGCGGAACCAGTCTCGCGGACTTGCTGTCCTCCGCGTTGCCCGACAAGAAAGATCGCTCTACAACCCCTTCCGGTCCTCTCACAAATGGCGCCGGTGACGGATCTGAATGCGCGTCCACGTCCAGCGGGGGATCTGGTCCGCAGGCTGCGCGTGCTCGGCCTCTGCCTTTCGATACCGCCGTGCTGCAGGAGGTGGTCGGCTGTGCACTTGACGATCTGCACCTCGATGACCCCATCAATCTTGTCGCGGCTCTGGACCGCGATCTCTCGGACGGCGAGGGCGCGCTCGCGGGCTCCGCTCCCGTCAACATTCCGTCGGCGCGACCCGCCCTGCGCGGGTTCAGTCCCCCGGGCGGCTCACCGCTGCCGGCTTTCCTGCGATACGGACCCGACGCGTTCGGTCTCAAGGCTGGCAGCTTCGGCGCGTTCGAGGGCGGCGGCTGCGAGGTGTCGCGGCTGCGCGAGGAGGTGGCGGCGGCGCGGGCGGCGGTGGCGCGGTGGGACGAGCGCGTGGCGCAGGCGCGGGCGGCGTGCGAGGCGTGGCAGCGCGAGGCGGAGGACGCGGCGCGCAAGGCGGCGGTGGCGGAGCGGCAGCGCGACGAGGCGCTGGCACACGCGCTGGCGCTGCGGCGCGAGCTGGACGCGCTGGCGCCGCGCCGGGACCCGCGGGCGCTGTCGCTGCCGGCGCTCAAGGCGCTGCAGTCGCGCGCGCGGGCGGACCTCGAGGAGATCGAGAAGGCGTTGTACCTGGAGACGGCCACCAAGTGCATGGGCTGCGAGGAGCAGCCCCGCAGCGTCACGCTGGCGCCGTGCAACCACTACGTGCTGTGCGACAAGTGCGCGGGCACCACGGACCACTGCCCCTACTGCCAGACCCCGGTGCAGAGACACCAATAG
Protein
MPSESKPLLTAQTEKPNHYKYLKEFRVEQCPSFLQHKCTQHRPFTCFHWHFNNQRRRRPVRKRDGTFNYSADNYCTKYNETSGVCEDGDECPFLHRTAGDTERRYHLRYYKTCMCVHDTDTRGLCTKNGAHCAFAHGAPDLRPPVLDVRELHALDADDDAAPNALDRERNLINEDPKWQDTNYVLSSYKTEPCKRPPRLCRQGYACPQYHNSKDKRRSPRKYKYRSTPCPNVKHGEEWGEPSNCEAGDTCGYCHTRTEQQFHPEIYKSTKCNDVQQAGYCPRGLFCAFAHVEPEEPGGREPGPGDCGTSLADLLSSALPDKKDRSTTPSGPLTNGAGDGSECASTSSGGSGPQAARARPLPFDTAVLQEVVGCALDDLHLDDPINLVAALDRDLSDGEGALAGSAPVNIPSARPALRGFSPPGGSPLPAFLRYGPDAFGLKAGSFGAFEGGGCEVSRLREEVAAARAAVARWDERVAQARAACEAWQREAEDAARKAAVAERQRDEALAHALALRRELDALAPRRDPRALSLPALKALQSRARADLEEIEKALYLETATKCMGCEEQPRSVTLAPCNHYVLCDKCAGTTDHCPYCQTPVQRHQ
Summary
Uniprot
Pubmed
EMBL
KQ459232
KPJ02599.1
RSAL01000145
RVE45925.1
KQ461155
KPJ08333.1
+ More
AGBW02008375 OWR53555.1 CU367882 CBH09250.1 KC469893 AGC92674.1 NWSH01001193 PCG72183.1 PCG72182.1 GBYB01002818 JAG72585.1 DS235179 EEB13021.1 PYGN01001553 PSN34114.1 APGK01039007 KB740966 KB632186 ENN76943.1 ERL89713.1 JRES01000409 KNC31529.1 CH964272 KRG00168.1 CM000070 KRT00031.1 CM000160 KRK04116.1 CH940650 KRF83760.1 CH933806 KRG01311.1
AGBW02008375 OWR53555.1 CU367882 CBH09250.1 KC469893 AGC92674.1 NWSH01001193 PCG72183.1 PCG72182.1 GBYB01002818 JAG72585.1 DS235179 EEB13021.1 PYGN01001553 PSN34114.1 APGK01039007 KB740966 KB632186 ENN76943.1 ERL89713.1 JRES01000409 KNC31529.1 CH964272 KRG00168.1 CM000070 KRT00031.1 CM000160 KRK04116.1 CH940650 KRF83760.1 CH933806 KRG01311.1
Proteomes
Interpro
SUPFAM
SSF90229
SSF90229
Gene 3D
CDD
ProteinModelPortal
PDB
5ELH
E-value=3.91578e-54,
Score=537
Ontologies
GO
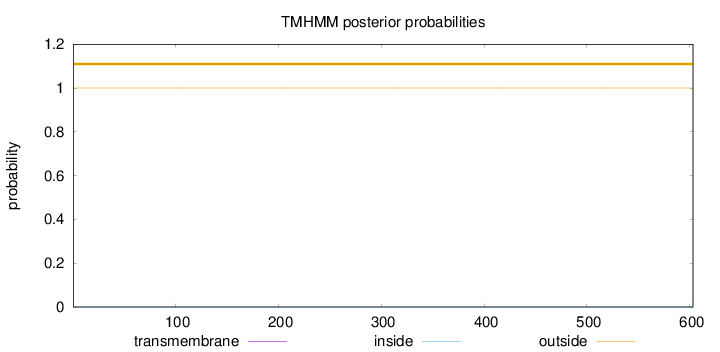
Topology
Length:
603
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8e-05
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00015
outside
1 - 603
Population Genetic Test Statistics
Pi
135.168496
Theta
172.547563
Tajima's D
-0.664624
CLR
0.429555
CSRT
0.210839458027099
Interpretation
Uncertain
