Gene
KWMTBOMO10098
Pre Gene Modal
BGIBMGA005549
Annotation
PREDICTED:_uncharacterized_protein_LOC101743057_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.083 PlasmaMembrane Reliability : 1.464
Sequence
CDS
ATGCGCGAGTACGTGTCGCGCGTGTGCGGCGCCGTGCGGGCCAGCGACGCGCTGCGGGCGCTGGGCCCCGGCCTGCCCGAGCTGCTGGTGGCGCAGCTGCGGGCGCATCTCGCGCTGCACAGAGCCGAAGAAGGCGTCCGCAAAAAGATCGAGGCGGGTCTTGCGGCGGCCGAGGCGAGGGTCCGCGCCTCCCACCCGATCCTGTCCCGGGCCGACTCGTGGAGGAACGAGAAGCTGGCGGCGGCGGCGCGGCGGCTCCGGGACGAGTGCCGCTGGACCGCGCTCGAGGACGCCGCCGCCGCCATGCAGGCGCACAAGCTGCACCAGCACCGCTACTTCCTGCTGCGGGACCTGGCCTTCCAGCGGGACCGGGAGCCCTTGTTGATGAAGGAGCTGCGCGCGGCGCGGACGGCGCGGCGCTCGTGGCAGTGGGCGCGGCGCGTGTGGCGGCCGGCGCGCTGGGCCGTGGTGCGACACTGCCGCGGCCGCGCCGAGCGCGTGCGCACCGCCGTGGCCGCCCGCGCGCCCGCCGCCCCGCCCCGCGCCCGCCCGCCCGCCGCGCCGCCAGTGTTCCTGCTGCGGAGGGACGCCGTCTCCACCACCTCCACGCGCTGGCCGGCCTGGAGACTGCTCAACCTCGCCTATAGAATATGGTGCTGGAGCTGGAACCTGGTGTACGTGCTGGGCGTGGTGGTGCCGTGGTGCTCCCCGCTGTCGCTGCGAGCGCTGGTCTGCGTCAAGCCGTACACGCCGGAGCTAGAGGTCTCCCAGGTGAATGGAGTTTTATATCCGAAGAAGAGCAGCGAAACCCACACGATGTGCTCGCGACTGCTGGCGCTGTGGAGGCACGTGTCCAAACAGCGCACGAAGTTCGAAACCGAACCGGACACCGGCCTACTGGGCAAGGGCTTGAGTCGGTGGGCGCACCGCGTGTGGTGGTACGGCGTGGTGGGGGGCGGCGGGTCGGCGGCGCTGCTGGTGGTGTTCCCGGTGTGCGCGCTGGCGGTGTCGGCGGCGGCGGTGCTGCTGGCGCTGGCGGCCCCGCTGTGGACGCCGCCGCTGGCGCTGGCCGCGCACGTGGCGCACGCCGTGCTCTACGACGCCGACTCGCCCGACCCAGCACTCAACAGGTGGTGTCCGCTGCTGGAGGTGGTGCTGTGGCGAGTAGCGGTGCTGGGCGTGCTGCAGCCCGCCGCCGCGCTGCTCACGGCGCTGCTGCTCTGTCCGCTGGCTGCTGCTGTCCTGCTCGTCGGTGAGCACATCCACCCCCCCTCCCCACCTTCTTCTTATCGGCCCACAGACGTCAACTGCTGGACATAG
Protein
MREYVSRVCGAVRASDALRALGPGLPELLVAQLRAHLALHRAEEGVRKKIEAGLAAAEARVRASHPILSRADSWRNEKLAAAARRLRDECRWTALEDAAAAMQAHKLHQHRYFLLRDLAFQRDREPLLMKELRAARTARRSWQWARRVWRPARWAVVRHCRGRAERVRTAVAARAPAAPPRARPPAAPPVFLLRRDAVSTTSTRWPAWRLLNLAYRIWCWSWNLVYVLGVVVPWCSPLSLRALVCVKPYTPELEVSQVNGVLYPKKSSETHTMCSRLLALWRHVSKQRTKFETEPDTGLLGKGLSRWAHRVWWYGVVGGGGSAALLVVFPVCALAVSAAAVLLALAAPLWTPPLALAAHVAHAVLYDADSPDPALNRWCPLLEVVLWRVAVLGVLQPAAALLTALLLCPLAAAVLLVGEHIHPPSPPSSYRPTDVNCWT
Summary
Uniprot
C9S269
L7X1E1
A0A3S2TH67
A0A194QAN7
A0A1Y1LRG3
A0A0N8P1T8
+ More
A0A2J7PKN4 A0A2J7PKN5 A0A1B6GED7 Q16JA6 B3M1R3 B4N977 A0A0Q9WTT3 A0A1S4FYV6 A0A034VDR0 A0A182H448 A0A0A1XHZ6 A0A0L0CL78 A0A034V9D0 A0A1I8P7K5 A0A1Y1LWM1 A0A1I8N8E1 A0A0K8W7H3 A0A2M3Z6U5 A0A0K8UAN3 A0A2M4AAR4 I5APC4 B4GF79 Q296F9 A0A3B0KKR6 A0A2M4AA66 A0A2M4A9I0 A0A2M4A9H2 A0A1B6D278 W5JSX9 A0A2M4BA49 A0A182FAG4 A0A182JKT9 A0A0M4EEX1 A0A182SVR5 A0A1I8N8E8 E0VI12 B4JHU8 B4KD88 A0A1Y1LT12 A0A154PK19 E2BMC9 A0A182TYD2 A0A151I699 A0A1B0CVD0 A0A182Y8Z6 A0A158P198 A0A151XHL2 N6SZE2 A0A151JX63 A0A195DKZ7 A0A1B0FDA1 A0A1Q3FFF4 A0A1A9Z8G2 A0A1B0AXJ2 A0A1Q3FFC0 A0A1A9VB52 A0A1A9XTJ4 A0A336M682 A0A182PLI3 A0A1L8D9Q9 A0A182W395 B0WG09 A0A182XF79 A0A182VCM5 A0A182RI14 A0A2A3EAL4 F4WUL8 A0A182HVQ0 A0A182KZR9 E2AWV3 K7IYD8 A0A232EMT0 A0A3Q0JG51 A0A0L7REM8 A0A182JP30 A0A0J7KVG7 Q7QA60 D6WZ19 A0A088AJY2 A0A0K8UGX8 A0A0K8WM17 A0A182MHJ3 A0A1Q3FFG6 A0A0C9R4I4 A0A0C9QQB0 A0A182QS17 A0A182NCW1 A0A151IGJ7 A0A1Q3FFJ5 A0A212FIJ4 A0A1A9X1V8 A0A2S2PIS0
A0A2J7PKN4 A0A2J7PKN5 A0A1B6GED7 Q16JA6 B3M1R3 B4N977 A0A0Q9WTT3 A0A1S4FYV6 A0A034VDR0 A0A182H448 A0A0A1XHZ6 A0A0L0CL78 A0A034V9D0 A0A1I8P7K5 A0A1Y1LWM1 A0A1I8N8E1 A0A0K8W7H3 A0A2M3Z6U5 A0A0K8UAN3 A0A2M4AAR4 I5APC4 B4GF79 Q296F9 A0A3B0KKR6 A0A2M4AA66 A0A2M4A9I0 A0A2M4A9H2 A0A1B6D278 W5JSX9 A0A2M4BA49 A0A182FAG4 A0A182JKT9 A0A0M4EEX1 A0A182SVR5 A0A1I8N8E8 E0VI12 B4JHU8 B4KD88 A0A1Y1LT12 A0A154PK19 E2BMC9 A0A182TYD2 A0A151I699 A0A1B0CVD0 A0A182Y8Z6 A0A158P198 A0A151XHL2 N6SZE2 A0A151JX63 A0A195DKZ7 A0A1B0FDA1 A0A1Q3FFF4 A0A1A9Z8G2 A0A1B0AXJ2 A0A1Q3FFC0 A0A1A9VB52 A0A1A9XTJ4 A0A336M682 A0A182PLI3 A0A1L8D9Q9 A0A182W395 B0WG09 A0A182XF79 A0A182VCM5 A0A182RI14 A0A2A3EAL4 F4WUL8 A0A182HVQ0 A0A182KZR9 E2AWV3 K7IYD8 A0A232EMT0 A0A3Q0JG51 A0A0L7REM8 A0A182JP30 A0A0J7KVG7 Q7QA60 D6WZ19 A0A088AJY2 A0A0K8UGX8 A0A0K8WM17 A0A182MHJ3 A0A1Q3FFG6 A0A0C9R4I4 A0A0C9QQB0 A0A182QS17 A0A182NCW1 A0A151IGJ7 A0A1Q3FFJ5 A0A212FIJ4 A0A1A9X1V8 A0A2S2PIS0
Pubmed
EMBL
CT955980
CBH09249.1
KC469893
AGC92671.2
RSAL01000145
RVE45926.1
+ More
KQ459232 KPJ02598.1 GEZM01050708 JAV75428.1 CH902617 KPU80844.1 NEVH01024535 PNF16892.1 PNF16890.1 GECZ01008993 JAS60776.1 CH478020 EAT34353.1 EDV44393.1 KPU80845.1 CH964232 EDW81624.1 KRF99500.1 GAKP01019029 GAKP01019027 JAC39925.1 JXUM01108785 JXUM01108786 JXUM01108787 GBXI01013390 GBXI01003691 JAD00902.1 JAD10601.1 JRES01000249 KNC32987.1 GAKP01019031 JAC39921.1 GEZM01050711 GEZM01050709 JAV75427.1 GDHF01018942 GDHF01005287 JAI33372.1 JAI47027.1 GGFM01003459 MBW24210.1 GDHF01028661 GDHF01003331 JAI23653.1 JAI48983.1 GGFK01004546 MBW37867.1 CM000070 EIM52809.1 CH479182 EDW34264.1 EAL28498.2 OUUW01000008 SPP84378.1 GGFK01004330 MBW37651.1 GGFK01004104 MBW37425.1 GGFK01004094 MBW37415.1 GEDC01017518 GEDC01017494 GEDC01010789 JAS19780.1 JAS19804.1 JAS26509.1 ADMH02000180 ETN67472.1 GGFJ01000749 MBW49890.1 CP012526 ALC46616.1 DS235179 EEB13018.1 CH916369 EDV92856.1 CH933806 EDW14870.2 GEZM01050707 JAV75430.1 KQ434931 KZC11804.1 GL449185 EFN83158.1 KQ976402 KYM92296.1 AJWK01030433 AJWK01030434 ADTU01006152 ADTU01006153 ADTU01006154 KQ982138 KYQ59700.1 APGK01058767 KB741291 ENN70608.1 KQ981619 KYN39251.1 KQ980762 KYN13560.1 CCAG010014354 GFDL01008769 JAV26276.1 JXJN01005276 GFDL01008805 JAV26240.1 UFQS01000590 UFQT01000590 SSX05179.1 SSX25540.1 GFDF01010987 JAV03097.1 DS231922 EDS26650.1 KZ288308 PBC28730.1 GL888373 EGI62099.1 APCN01005809 GL443425 EFN62073.1 NNAY01003297 OXU19664.1 KQ414608 KOC69407.1 LBMM01002835 KMQ94296.1 AAAB01008898 EAA09220.5 KQ971363 EFA09040.2 GDHF01026498 JAI25816.1 GDHF01000111 JAI52203.1 AXCM01004942 GFDL01008748 JAV26297.1 GBYB01002819 GBYB01002820 JAG72586.1 JAG72587.1 GBYB01002817 JAG72584.1 AXCN02001559 KQ977743 KYN00183.1 GFDL01008763 JAV26282.1 AGBW02008375 OWR53556.1 GGMR01016720 MBY29339.1
KQ459232 KPJ02598.1 GEZM01050708 JAV75428.1 CH902617 KPU80844.1 NEVH01024535 PNF16892.1 PNF16890.1 GECZ01008993 JAS60776.1 CH478020 EAT34353.1 EDV44393.1 KPU80845.1 CH964232 EDW81624.1 KRF99500.1 GAKP01019029 GAKP01019027 JAC39925.1 JXUM01108785 JXUM01108786 JXUM01108787 GBXI01013390 GBXI01003691 JAD00902.1 JAD10601.1 JRES01000249 KNC32987.1 GAKP01019031 JAC39921.1 GEZM01050711 GEZM01050709 JAV75427.1 GDHF01018942 GDHF01005287 JAI33372.1 JAI47027.1 GGFM01003459 MBW24210.1 GDHF01028661 GDHF01003331 JAI23653.1 JAI48983.1 GGFK01004546 MBW37867.1 CM000070 EIM52809.1 CH479182 EDW34264.1 EAL28498.2 OUUW01000008 SPP84378.1 GGFK01004330 MBW37651.1 GGFK01004104 MBW37425.1 GGFK01004094 MBW37415.1 GEDC01017518 GEDC01017494 GEDC01010789 JAS19780.1 JAS19804.1 JAS26509.1 ADMH02000180 ETN67472.1 GGFJ01000749 MBW49890.1 CP012526 ALC46616.1 DS235179 EEB13018.1 CH916369 EDV92856.1 CH933806 EDW14870.2 GEZM01050707 JAV75430.1 KQ434931 KZC11804.1 GL449185 EFN83158.1 KQ976402 KYM92296.1 AJWK01030433 AJWK01030434 ADTU01006152 ADTU01006153 ADTU01006154 KQ982138 KYQ59700.1 APGK01058767 KB741291 ENN70608.1 KQ981619 KYN39251.1 KQ980762 KYN13560.1 CCAG010014354 GFDL01008769 JAV26276.1 JXJN01005276 GFDL01008805 JAV26240.1 UFQS01000590 UFQT01000590 SSX05179.1 SSX25540.1 GFDF01010987 JAV03097.1 DS231922 EDS26650.1 KZ288308 PBC28730.1 GL888373 EGI62099.1 APCN01005809 GL443425 EFN62073.1 NNAY01003297 OXU19664.1 KQ414608 KOC69407.1 LBMM01002835 KMQ94296.1 AAAB01008898 EAA09220.5 KQ971363 EFA09040.2 GDHF01026498 JAI25816.1 GDHF01000111 JAI52203.1 AXCM01004942 GFDL01008748 JAV26297.1 GBYB01002819 GBYB01002820 JAG72586.1 JAG72587.1 GBYB01002817 JAG72584.1 AXCN02001559 KQ977743 KYN00183.1 GFDL01008763 JAV26282.1 AGBW02008375 OWR53556.1 GGMR01016720 MBY29339.1
Proteomes
UP000283053
UP000053268
UP000007801
UP000235965
UP000008820
UP000007798
+ More
UP000069940 UP000037069 UP000095300 UP000095301 UP000001819 UP000008744 UP000268350 UP000000673 UP000069272 UP000075880 UP000092553 UP000075901 UP000009046 UP000001070 UP000009192 UP000076502 UP000008237 UP000075902 UP000078540 UP000092461 UP000076408 UP000005205 UP000075809 UP000019118 UP000078541 UP000078492 UP000092444 UP000092445 UP000092460 UP000078200 UP000092443 UP000075885 UP000075920 UP000002320 UP000076407 UP000075903 UP000075900 UP000242457 UP000007755 UP000075840 UP000075882 UP000000311 UP000002358 UP000215335 UP000079169 UP000053825 UP000075881 UP000036403 UP000007062 UP000007266 UP000005203 UP000075883 UP000075886 UP000075884 UP000078542 UP000007151 UP000091820
UP000069940 UP000037069 UP000095300 UP000095301 UP000001819 UP000008744 UP000268350 UP000000673 UP000069272 UP000075880 UP000092553 UP000075901 UP000009046 UP000001070 UP000009192 UP000076502 UP000008237 UP000075902 UP000078540 UP000092461 UP000076408 UP000005205 UP000075809 UP000019118 UP000078541 UP000078492 UP000092444 UP000092445 UP000092460 UP000078200 UP000092443 UP000075885 UP000075920 UP000002320 UP000076407 UP000075903 UP000075900 UP000242457 UP000007755 UP000075840 UP000075882 UP000000311 UP000002358 UP000215335 UP000079169 UP000053825 UP000075881 UP000036403 UP000007062 UP000007266 UP000005203 UP000075883 UP000075886 UP000075884 UP000078542 UP000007151 UP000091820
Interpro
IPR036640
ABC1_TM_sf
SUPFAM
SSF90123
SSF90123
ProteinModelPortal
C9S269
L7X1E1
A0A3S2TH67
A0A194QAN7
A0A1Y1LRG3
A0A0N8P1T8
+ More
A0A2J7PKN4 A0A2J7PKN5 A0A1B6GED7 Q16JA6 B3M1R3 B4N977 A0A0Q9WTT3 A0A1S4FYV6 A0A034VDR0 A0A182H448 A0A0A1XHZ6 A0A0L0CL78 A0A034V9D0 A0A1I8P7K5 A0A1Y1LWM1 A0A1I8N8E1 A0A0K8W7H3 A0A2M3Z6U5 A0A0K8UAN3 A0A2M4AAR4 I5APC4 B4GF79 Q296F9 A0A3B0KKR6 A0A2M4AA66 A0A2M4A9I0 A0A2M4A9H2 A0A1B6D278 W5JSX9 A0A2M4BA49 A0A182FAG4 A0A182JKT9 A0A0M4EEX1 A0A182SVR5 A0A1I8N8E8 E0VI12 B4JHU8 B4KD88 A0A1Y1LT12 A0A154PK19 E2BMC9 A0A182TYD2 A0A151I699 A0A1B0CVD0 A0A182Y8Z6 A0A158P198 A0A151XHL2 N6SZE2 A0A151JX63 A0A195DKZ7 A0A1B0FDA1 A0A1Q3FFF4 A0A1A9Z8G2 A0A1B0AXJ2 A0A1Q3FFC0 A0A1A9VB52 A0A1A9XTJ4 A0A336M682 A0A182PLI3 A0A1L8D9Q9 A0A182W395 B0WG09 A0A182XF79 A0A182VCM5 A0A182RI14 A0A2A3EAL4 F4WUL8 A0A182HVQ0 A0A182KZR9 E2AWV3 K7IYD8 A0A232EMT0 A0A3Q0JG51 A0A0L7REM8 A0A182JP30 A0A0J7KVG7 Q7QA60 D6WZ19 A0A088AJY2 A0A0K8UGX8 A0A0K8WM17 A0A182MHJ3 A0A1Q3FFG6 A0A0C9R4I4 A0A0C9QQB0 A0A182QS17 A0A182NCW1 A0A151IGJ7 A0A1Q3FFJ5 A0A212FIJ4 A0A1A9X1V8 A0A2S2PIS0
A0A2J7PKN4 A0A2J7PKN5 A0A1B6GED7 Q16JA6 B3M1R3 B4N977 A0A0Q9WTT3 A0A1S4FYV6 A0A034VDR0 A0A182H448 A0A0A1XHZ6 A0A0L0CL78 A0A034V9D0 A0A1I8P7K5 A0A1Y1LWM1 A0A1I8N8E1 A0A0K8W7H3 A0A2M3Z6U5 A0A0K8UAN3 A0A2M4AAR4 I5APC4 B4GF79 Q296F9 A0A3B0KKR6 A0A2M4AA66 A0A2M4A9I0 A0A2M4A9H2 A0A1B6D278 W5JSX9 A0A2M4BA49 A0A182FAG4 A0A182JKT9 A0A0M4EEX1 A0A182SVR5 A0A1I8N8E8 E0VI12 B4JHU8 B4KD88 A0A1Y1LT12 A0A154PK19 E2BMC9 A0A182TYD2 A0A151I699 A0A1B0CVD0 A0A182Y8Z6 A0A158P198 A0A151XHL2 N6SZE2 A0A151JX63 A0A195DKZ7 A0A1B0FDA1 A0A1Q3FFF4 A0A1A9Z8G2 A0A1B0AXJ2 A0A1Q3FFC0 A0A1A9VB52 A0A1A9XTJ4 A0A336M682 A0A182PLI3 A0A1L8D9Q9 A0A182W395 B0WG09 A0A182XF79 A0A182VCM5 A0A182RI14 A0A2A3EAL4 F4WUL8 A0A182HVQ0 A0A182KZR9 E2AWV3 K7IYD8 A0A232EMT0 A0A3Q0JG51 A0A0L7REM8 A0A182JP30 A0A0J7KVG7 Q7QA60 D6WZ19 A0A088AJY2 A0A0K8UGX8 A0A0K8WM17 A0A182MHJ3 A0A1Q3FFG6 A0A0C9R4I4 A0A0C9QQB0 A0A182QS17 A0A182NCW1 A0A151IGJ7 A0A1Q3FFJ5 A0A212FIJ4 A0A1A9X1V8 A0A2S2PIS0
Ontologies
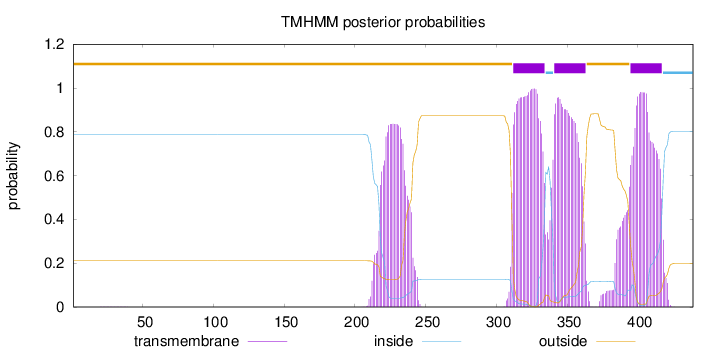
Topology
Length:
439
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
85.77117
Exp number, first 60 AAs:
0.00541
Total prob of N-in:
0.78915
outside
1 - 311
TMhelix
312 - 334
inside
335 - 340
TMhelix
341 - 363
outside
364 - 394
TMhelix
395 - 417
inside
418 - 439
Population Genetic Test Statistics
Pi
149.145432
Theta
145.311852
Tajima's D
-0.924266
CLR
10.731074
CSRT
0.153042347882606
Interpretation
Uncertain
