Gene
KWMTBOMO10097
Pre Gene Modal
BGIBMGA005549
Annotation
PREDICTED:_uncharacterized_protein_LOC101743057_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.106 Mitochondrial Reliability : 1.747
Sequence
CDS
ATGTCAGGAGATCGAGCAACGCGGCCGAACTCTTTAGTGTTGGATACGCCGGCACCGGAGCGCGAACCTCTGCGCTTCGATGTGCAGTTAGTTGGAGCCCCACCGGAGGTTGAGCAGCTGGTTAATAACATCAAACAGGTAGCCGAAGAATTCTTGTATCACTGGAAAACTTTTCCCATTGTATTACCTCCAACACGGTTTGGTCCCGGTAACCGTCCATGTGACATCATTATTCCACCTCCTTGCGATGAGGTGGATGCAGCTGCTCTAGATGCTGGACCGGAACCGCAACCTCTGTCGCCCAAACAACTACAATCTATTAGGGAGAAAGGAGAGTTCGAAGTACCTTCCCGTCACTTTCCTGGTCAAACGCACGTGTGGCGAGTGGCGGGCTGGTTGCAGCGCGGTAAGGTCCGAGCCCAAGAGGAACTCTACCGACACGTGGCGAGGGCGTTGGCGCTGATCGTGGTAGTCGCCAGGGACCGACTTGTCAAAGATGAACCGCTTTCTGTACGAGCGGGGATCAAATCGCTTCTGGCCGGCTTGCACAGGTTATTGGATCTGGCCATTGGGATGCCATCAATGGAGGCCCGGGATCTCGATAAGAAGATACGCGAGGAAAGATCACGATATCTGGTGGCAGAGTTGATATGCAAGCCGGAGCAACAGGAGGACGTGACGGCGTTGGCGGGCTGGGTGTGGCGAGCCTTACGCCGCGCCGCCGCAGAGAAGTGCGCCCCCCGCTGCCCCCCGCCCGTCCCCACGCTCTACACCACGCCCCAACACACGCAGGTTGACCTGCGTCTGTACCGTCGCGAGGTGATGAGGAGTGCCCTGGGTCCGCTGCAGGCCGTGCTGGAGCGCGAGGCCCGCGGCTGGTTCCTGCACTTCCGGGAACAACAGGCCGCCCATCTCGCGCGGGAGAAGATGCCCCTGAAGGATATCGAGGAGGTCGGTCAATGA
Protein
MSGDRATRPNSLVLDTPAPEREPLRFDVQLVGAPPEVEQLVNNIKQVAEEFLYHWKTFPIVLPPTRFGPGNRPCDIIIPPPCDEVDAAALDAGPEPQPLSPKQLQSIREKGEFEVPSRHFPGQTHVWRVAGWLQRGKVRAQEELYRHVARALALIVVVARDRLVKDEPLSVRAGIKSLLAGLHRLLDLAIGMPSMEARDLDKKIREERSRYLVAELICKPEQQEDVTALAGWVWRALRRAAAEKCAPRCPPPVPTLYTTPQHTQVDLRLYRREVMRSALGPLQAVLEREARGWFLHFREQQAAHLAREKMPLKDIEEVGQ
Summary
Uniprot
H9J7Q7
A0A212EMI5
A0A194QTK6
A0A194QAN7
A0A2A4J2V5
C9S269
+ More
L7X1E1 A0A1B6L0C9 A0A2A4J3T3 A0A1B6JLT9 A0A3S2TH67 V5GYT4 D6WZ19 A0A0A9VR70 A0A1W4X2F4 T1H7N8 Q7KTS3 A0A1W4UWR5 A0A0K8UGX8 I5APC4 A0A146KR81 A0A224XH11 A0A0R1E0T9 A0A069DY18 N6SZE2 A0A0N8P1T8 W8AR39 A0A0K8W7H3 A0A034VDR0 A0A0C9QQB0 A0A0A1XHZ6 A0A0Q9WTT3 A0A1W4X2B1 A0A1W4XCV9 A0A1I8P7K5 A0A1Y1LWM1 A0A0M4EEX1 A0A1I8N8E1 B4I417 A0A2S2PPY6 A0A1A9VB52 B4QWX0 Q9VNC9 V9I920 A0A1W4UWG2 B4GF79 A0A336M682 A0A2J7PKN4 Q296F9 A0A1A9Z8G2 A0A2S2QUX5 A0A1B0FDA1 B3P2E8 A0A0K8WM17 A0A1B0AXJ2 B4PVB3 A0A1A9XTJ4 A0A182JKT9 Q16JA6 A0A182H448 A0A3B0KKR6 B3M1R3 U4TYG0 W8AJ43 A0A0K8UAN3 A0A034V9D0 B4N977 A0A0C9R4I4 J9JXH7 A0A151JX63 E2AWV3 A0A195DKZ7 B4KD88 A0A1B6D278 A0A151IGJ7 E2BMC9 A0A158P198 A0A182QS17 B4LZB3 A0A182JP30 A0A151I699 A0A0L0CL78 A0A1I8N8E8 B4JHU8 A0A026W1X6 A0A084WPA7 W4VR24 A0A2J7PKN5 V9IA00 F4WUL8 A0A1J1IIY0 A0A182NCW1 A0A310SH47 A0A182KZR9 Q7QA60 K7IYD8 A0A182PLI3 A0A2A3EAL4 A0A088AJY2
L7X1E1 A0A1B6L0C9 A0A2A4J3T3 A0A1B6JLT9 A0A3S2TH67 V5GYT4 D6WZ19 A0A0A9VR70 A0A1W4X2F4 T1H7N8 Q7KTS3 A0A1W4UWR5 A0A0K8UGX8 I5APC4 A0A146KR81 A0A224XH11 A0A0R1E0T9 A0A069DY18 N6SZE2 A0A0N8P1T8 W8AR39 A0A0K8W7H3 A0A034VDR0 A0A0C9QQB0 A0A0A1XHZ6 A0A0Q9WTT3 A0A1W4X2B1 A0A1W4XCV9 A0A1I8P7K5 A0A1Y1LWM1 A0A0M4EEX1 A0A1I8N8E1 B4I417 A0A2S2PPY6 A0A1A9VB52 B4QWX0 Q9VNC9 V9I920 A0A1W4UWG2 B4GF79 A0A336M682 A0A2J7PKN4 Q296F9 A0A1A9Z8G2 A0A2S2QUX5 A0A1B0FDA1 B3P2E8 A0A0K8WM17 A0A1B0AXJ2 B4PVB3 A0A1A9XTJ4 A0A182JKT9 Q16JA6 A0A182H448 A0A3B0KKR6 B3M1R3 U4TYG0 W8AJ43 A0A0K8UAN3 A0A034V9D0 B4N977 A0A0C9R4I4 J9JXH7 A0A151JX63 E2AWV3 A0A195DKZ7 B4KD88 A0A1B6D278 A0A151IGJ7 E2BMC9 A0A158P198 A0A182QS17 B4LZB3 A0A182JP30 A0A151I699 A0A0L0CL78 A0A1I8N8E8 B4JHU8 A0A026W1X6 A0A084WPA7 W4VR24 A0A2J7PKN5 V9IA00 F4WUL8 A0A1J1IIY0 A0A182NCW1 A0A310SH47 A0A182KZR9 Q7QA60 K7IYD8 A0A182PLI3 A0A2A3EAL4 A0A088AJY2
Pubmed
19121390
22118469
26354079
23674305
18362917
19820115
+ More
25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17994087 26823975 17550304 26334808 23537049 24495485 25348373 25830018 28004739 25315136 17510324 26483478 18057021 20798317 21347285 26108605 24508170 30249741 24438588 21719571 20966253 12364791 14747013 17210077 20075255
25401762 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17994087 26823975 17550304 26334808 23537049 24495485 25348373 25830018 28004739 25315136 17510324 26483478 18057021 20798317 21347285 26108605 24508170 30249741 24438588 21719571 20966253 12364791 14747013 17210077 20075255
EMBL
BABH01020278
BABH01020279
BABH01020280
BABH01020281
BABH01020282
AGBW02013813
+ More
OWR42704.1 KQ461155 KPJ08335.1 KQ459232 KPJ02598.1 NWSH01003388 PCG66395.1 CT955980 CBH09249.1 KC469893 AGC92671.2 GEBQ01022814 JAT17163.1 PCG66396.1 GECU01007502 JAT00205.1 RSAL01000145 RVE45926.1 GALX01002858 JAB65608.1 KQ971363 EFA09040.2 GBHO01045335 JAF98268.1 ACPB03010566 AE014297 AAS65097.1 AGB95669.1 GDHF01026498 JAI25816.1 CM000070 EIM52809.1 GDHC01020887 GDHC01004853 JAP97741.1 JAQ13776.1 GFTR01008676 JAW07750.1 CM000160 KRK02719.1 GBGD01000163 JAC88726.1 APGK01058767 KB741291 ENN70608.1 CH902617 KPU80844.1 GAMC01018023 JAB88532.1 GDHF01018942 GDHF01005287 JAI33372.1 JAI47027.1 GAKP01019029 GAKP01019027 JAC39925.1 GBYB01002817 JAG72584.1 GBXI01013390 GBXI01003691 JAD00902.1 JAD10601.1 CH964232 KRF99500.1 GEZM01050711 GEZM01050709 JAV75427.1 CP012526 ALC46616.1 CH480821 EDW54960.1 GGMR01018785 MBY31404.1 CM000364 EDX11732.1 AY051864 AAF52014.2 AAK93288.1 JR036400 JR036401 AEY57126.1 AEY57127.1 CH479182 EDW34264.1 UFQS01000590 UFQT01000590 SSX05179.1 SSX25540.1 NEVH01024535 PNF16892.1 EAL28498.2 GGMS01011729 MBY80932.1 CCAG010014354 CH954181 EDV47898.1 GDHF01000111 JAI52203.1 JXJN01005276 EDW95782.1 CH478020 EAT34353.1 JXUM01108785 JXUM01108786 JXUM01108787 OUUW01000008 SPP84378.1 EDV44393.1 KPU80845.1 KB631759 ERL85852.1 GAMC01018025 JAB88530.1 GDHF01028661 GDHF01003331 JAI23653.1 JAI48983.1 GAKP01019031 JAC39921.1 EDW81624.1 GBYB01002819 GBYB01002820 JAG72586.1 JAG72587.1 ABLF02027144 KQ981619 KYN39251.1 GL443425 EFN62073.1 KQ980762 KYN13560.1 CH933806 EDW14870.2 GEDC01017518 GEDC01017494 GEDC01010789 JAS19780.1 JAS19804.1 JAS26509.1 KQ977743 KYN00183.1 GL449185 EFN83158.1 ADTU01006152 ADTU01006153 ADTU01006154 AXCN02001559 CH940650 EDW68148.2 KQ976402 KYM92296.1 JRES01000249 KNC32987.1 CH916369 EDV92856.1 KK107503 QOIP01000006 EZA49576.1 RLU21832.1 ATLV01025005 ATLV01025006 ATLV01025007 KE525367 KFB52051.1 GANO01004909 JAB54962.1 PNF16890.1 JR036397 JR036398 AEY57124.1 AEY57125.1 GL888373 EGI62099.1 CVRI01000048 CRK99022.1 KQ763581 OAD54959.1 AAAB01008898 EAA09220.5 KZ288308 PBC28730.1
OWR42704.1 KQ461155 KPJ08335.1 KQ459232 KPJ02598.1 NWSH01003388 PCG66395.1 CT955980 CBH09249.1 KC469893 AGC92671.2 GEBQ01022814 JAT17163.1 PCG66396.1 GECU01007502 JAT00205.1 RSAL01000145 RVE45926.1 GALX01002858 JAB65608.1 KQ971363 EFA09040.2 GBHO01045335 JAF98268.1 ACPB03010566 AE014297 AAS65097.1 AGB95669.1 GDHF01026498 JAI25816.1 CM000070 EIM52809.1 GDHC01020887 GDHC01004853 JAP97741.1 JAQ13776.1 GFTR01008676 JAW07750.1 CM000160 KRK02719.1 GBGD01000163 JAC88726.1 APGK01058767 KB741291 ENN70608.1 CH902617 KPU80844.1 GAMC01018023 JAB88532.1 GDHF01018942 GDHF01005287 JAI33372.1 JAI47027.1 GAKP01019029 GAKP01019027 JAC39925.1 GBYB01002817 JAG72584.1 GBXI01013390 GBXI01003691 JAD00902.1 JAD10601.1 CH964232 KRF99500.1 GEZM01050711 GEZM01050709 JAV75427.1 CP012526 ALC46616.1 CH480821 EDW54960.1 GGMR01018785 MBY31404.1 CM000364 EDX11732.1 AY051864 AAF52014.2 AAK93288.1 JR036400 JR036401 AEY57126.1 AEY57127.1 CH479182 EDW34264.1 UFQS01000590 UFQT01000590 SSX05179.1 SSX25540.1 NEVH01024535 PNF16892.1 EAL28498.2 GGMS01011729 MBY80932.1 CCAG010014354 CH954181 EDV47898.1 GDHF01000111 JAI52203.1 JXJN01005276 EDW95782.1 CH478020 EAT34353.1 JXUM01108785 JXUM01108786 JXUM01108787 OUUW01000008 SPP84378.1 EDV44393.1 KPU80845.1 KB631759 ERL85852.1 GAMC01018025 JAB88530.1 GDHF01028661 GDHF01003331 JAI23653.1 JAI48983.1 GAKP01019031 JAC39921.1 EDW81624.1 GBYB01002819 GBYB01002820 JAG72586.1 JAG72587.1 ABLF02027144 KQ981619 KYN39251.1 GL443425 EFN62073.1 KQ980762 KYN13560.1 CH933806 EDW14870.2 GEDC01017518 GEDC01017494 GEDC01010789 JAS19780.1 JAS19804.1 JAS26509.1 KQ977743 KYN00183.1 GL449185 EFN83158.1 ADTU01006152 ADTU01006153 ADTU01006154 AXCN02001559 CH940650 EDW68148.2 KQ976402 KYM92296.1 JRES01000249 KNC32987.1 CH916369 EDV92856.1 KK107503 QOIP01000006 EZA49576.1 RLU21832.1 ATLV01025005 ATLV01025006 ATLV01025007 KE525367 KFB52051.1 GANO01004909 JAB54962.1 PNF16890.1 JR036397 JR036398 AEY57124.1 AEY57125.1 GL888373 EGI62099.1 CVRI01000048 CRK99022.1 KQ763581 OAD54959.1 AAAB01008898 EAA09220.5 KZ288308 PBC28730.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000283053
+ More
UP000007266 UP000192223 UP000015103 UP000000803 UP000192221 UP000001819 UP000002282 UP000019118 UP000007801 UP000007798 UP000095300 UP000092553 UP000095301 UP000001292 UP000078200 UP000000304 UP000008744 UP000235965 UP000092445 UP000092444 UP000008711 UP000092460 UP000092443 UP000075880 UP000008820 UP000069940 UP000268350 UP000030742 UP000007819 UP000078541 UP000000311 UP000078492 UP000009192 UP000078542 UP000008237 UP000005205 UP000075886 UP000008792 UP000075881 UP000078540 UP000037069 UP000001070 UP000053097 UP000279307 UP000030765 UP000007755 UP000183832 UP000075884 UP000075882 UP000007062 UP000002358 UP000075885 UP000242457 UP000005203
UP000007266 UP000192223 UP000015103 UP000000803 UP000192221 UP000001819 UP000002282 UP000019118 UP000007801 UP000007798 UP000095300 UP000092553 UP000095301 UP000001292 UP000078200 UP000000304 UP000008744 UP000235965 UP000092445 UP000092444 UP000008711 UP000092460 UP000092443 UP000075880 UP000008820 UP000069940 UP000268350 UP000030742 UP000007819 UP000078541 UP000000311 UP000078492 UP000009192 UP000078542 UP000008237 UP000005205 UP000075886 UP000008792 UP000075881 UP000078540 UP000037069 UP000001070 UP000053097 UP000279307 UP000030765 UP000007755 UP000183832 UP000075884 UP000075882 UP000007062 UP000002358 UP000075885 UP000242457 UP000005203
Interpro
IPR036640
ABC1_TM_sf
SUPFAM
SSF90123
SSF90123
ProteinModelPortal
H9J7Q7
A0A212EMI5
A0A194QTK6
A0A194QAN7
A0A2A4J2V5
C9S269
+ More
L7X1E1 A0A1B6L0C9 A0A2A4J3T3 A0A1B6JLT9 A0A3S2TH67 V5GYT4 D6WZ19 A0A0A9VR70 A0A1W4X2F4 T1H7N8 Q7KTS3 A0A1W4UWR5 A0A0K8UGX8 I5APC4 A0A146KR81 A0A224XH11 A0A0R1E0T9 A0A069DY18 N6SZE2 A0A0N8P1T8 W8AR39 A0A0K8W7H3 A0A034VDR0 A0A0C9QQB0 A0A0A1XHZ6 A0A0Q9WTT3 A0A1W4X2B1 A0A1W4XCV9 A0A1I8P7K5 A0A1Y1LWM1 A0A0M4EEX1 A0A1I8N8E1 B4I417 A0A2S2PPY6 A0A1A9VB52 B4QWX0 Q9VNC9 V9I920 A0A1W4UWG2 B4GF79 A0A336M682 A0A2J7PKN4 Q296F9 A0A1A9Z8G2 A0A2S2QUX5 A0A1B0FDA1 B3P2E8 A0A0K8WM17 A0A1B0AXJ2 B4PVB3 A0A1A9XTJ4 A0A182JKT9 Q16JA6 A0A182H448 A0A3B0KKR6 B3M1R3 U4TYG0 W8AJ43 A0A0K8UAN3 A0A034V9D0 B4N977 A0A0C9R4I4 J9JXH7 A0A151JX63 E2AWV3 A0A195DKZ7 B4KD88 A0A1B6D278 A0A151IGJ7 E2BMC9 A0A158P198 A0A182QS17 B4LZB3 A0A182JP30 A0A151I699 A0A0L0CL78 A0A1I8N8E8 B4JHU8 A0A026W1X6 A0A084WPA7 W4VR24 A0A2J7PKN5 V9IA00 F4WUL8 A0A1J1IIY0 A0A182NCW1 A0A310SH47 A0A182KZR9 Q7QA60 K7IYD8 A0A182PLI3 A0A2A3EAL4 A0A088AJY2
L7X1E1 A0A1B6L0C9 A0A2A4J3T3 A0A1B6JLT9 A0A3S2TH67 V5GYT4 D6WZ19 A0A0A9VR70 A0A1W4X2F4 T1H7N8 Q7KTS3 A0A1W4UWR5 A0A0K8UGX8 I5APC4 A0A146KR81 A0A224XH11 A0A0R1E0T9 A0A069DY18 N6SZE2 A0A0N8P1T8 W8AR39 A0A0K8W7H3 A0A034VDR0 A0A0C9QQB0 A0A0A1XHZ6 A0A0Q9WTT3 A0A1W4X2B1 A0A1W4XCV9 A0A1I8P7K5 A0A1Y1LWM1 A0A0M4EEX1 A0A1I8N8E1 B4I417 A0A2S2PPY6 A0A1A9VB52 B4QWX0 Q9VNC9 V9I920 A0A1W4UWG2 B4GF79 A0A336M682 A0A2J7PKN4 Q296F9 A0A1A9Z8G2 A0A2S2QUX5 A0A1B0FDA1 B3P2E8 A0A0K8WM17 A0A1B0AXJ2 B4PVB3 A0A1A9XTJ4 A0A182JKT9 Q16JA6 A0A182H448 A0A3B0KKR6 B3M1R3 U4TYG0 W8AJ43 A0A0K8UAN3 A0A034V9D0 B4N977 A0A0C9R4I4 J9JXH7 A0A151JX63 E2AWV3 A0A195DKZ7 B4KD88 A0A1B6D278 A0A151IGJ7 E2BMC9 A0A158P198 A0A182QS17 B4LZB3 A0A182JP30 A0A151I699 A0A0L0CL78 A0A1I8N8E8 B4JHU8 A0A026W1X6 A0A084WPA7 W4VR24 A0A2J7PKN5 V9IA00 F4WUL8 A0A1J1IIY0 A0A182NCW1 A0A310SH47 A0A182KZR9 Q7QA60 K7IYD8 A0A182PLI3 A0A2A3EAL4 A0A088AJY2
Ontologies
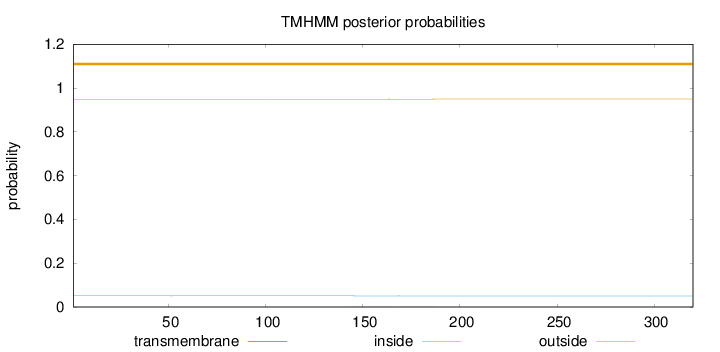
Topology
Length:
320
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01229
Exp number, first 60 AAs:
0.00088
Total prob of N-in:
0.05111
outside
1 - 320
Population Genetic Test Statistics
Pi
212.446335
Theta
119.735595
Tajima's D
0.089371
CLR
1.133123
CSRT
0.394380280985951
Interpretation
Uncertain
