Gene
KWMTBOMO10093 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005664
Annotation
trehalase_precursor_[Bombyx_mori]
Full name
Trehalase
Alternative Name
Alpha-trehalose glucohydrolase
Alpha,alpha-trehalase
Alpha,alpha-trehalose glucohydrolase
Alpha,alpha-trehalase
Alpha,alpha-trehalose glucohydrolase
Location in the cell
Cytoplasmic Reliability : 2.592
Sequence
CDS
ATGAGGTTGTTTCTACTACTGGTGGGATTGACGACGGTAATCGCGGATGACCTACCGCCGACTTGCATCAGACCAGTTTACTGCAACAGTACGCTGCTTCATTACGTTCAAATGGCCAGACTCTACCCCGATTCTAAAACTTTCGTCGATTTTCAAATGCGTAAAGACGAGAACGCAACACTGTCCGCCTTCCAAGAGCTGTTGGACCGCACGAATCATAATCCAACGAAAGAAGACCTGCAGGAATTTGTCGTTGACTTTTTTGATGAAACCAGCGAACTTGAAGAATGGAAACCGGATGACCATAAAGAAAACCCGCCTTTCCTCGCCAAAATTCGTGATGAAGGTTTCCGGGAATTCGCTAAAGCTTTAAATGATATTTGGCCAACTTTAGCCAGACGCGTTAAGCCCTCGGTCCTTGAAAAACCCGAACAATCGAGTCTCGTGCCTATGACACATGGCTTCATCGTTCCTGGAGGTCGATTTAAAGAAATCTACTATTGGGACGCATATTGGATTATCGAAGGTCTTCTAATCACTGACATGACCGAAACTGCAAAAGGAATGATCGAAAACCTAATCGAGCTTCTGTATAAATTTGGCCACATTCCCAATGGAAGCAGATGGTATTACCAAGAACGTAGCCAGCCTCCTTTATTGGCGGCCATGATAAAGCTTTATTACGAAAAGACTAAGGACATCGAATTCATCAGAAAGTATATTAGTGCTTTAGAAAAGGAATTAGAATATTGGCTGGACACTCATCTTATCGCTTTCAACAAGAACGACAGAGTTTACACTCTCCTGAGGTATTATATTCCAAGCGCTGGTCCTCGACCGGAGTCCTATTACGAAGATTACGAATTGGCTCAGAAATTAAACAAGACTACTGATCCTAATGATATTTACGCCGATTTAAAAAGCGCCGCTGAGAGCGGGTGGGACTTTTCTACGCGCTGGTTTATTTCAGAAAGCGGCGACAACAGTGGTAATCTAACCAATTTAAACACAAAGAACGTTATCCCCGTAGACTTGAATGCTATTTTCGCCGGAGCGTTACAGATTACGGCGAACTTCCAAGCTATATTAAAGAATCCTCGGAGAGCGGCTCACTGGGGCTACATGGCCGAACAATGGAGAAGTTCTATAGAGCAAGCGTTATGGGATGAAGAAGACGGAGTCTGGCACGACTACGACATATTGAATAATAAGCCACGTAGATATTTTTACACTAGCAACCTAGCTCCCCTGTGGATGAACGCAGTCGAAAAACCATTTCTAGCTAAGCATGGCGCTAGAGTCCTGGAGTACCTTCACGAATCGCAAGCGCTCGAGTACCCCGGAGGTGTCCCGGTGTCTCTAGTCAACAGTGGAGAACAATGGGACTTCCCTAACGCTTGGCCTCCAGAAGTCAGCATTGTCGTTACGGCTATCCAAAACATTGGTTCCGAGGAGAGTAGTAAATTGGCAAAAGAACTCGCCCAGGTCTGGGTGAGAGCTTGTAAATCAGGATTTACCGAAAAGAAACAGATGTTTGAGAAGTACGATGCGTTGAACGCTGGTAAATACGGCGGCGGCGGTGAATACACGGTTCAAGATGGATTCGGATGGTCGAACGGCGTTGTCCTGGAGTTCTTAGATAGGTACGGAGCTGTACTGACTTCGGTGGACTCTGTAGATGCTAGTGCAAATAACGGTCAGTCAAACGAAGAATCTGAGACGGATTCTAAGGAAAAATAA
Protein
MRLFLLLVGLTTVIADDLPPTCIRPVYCNSTLLHYVQMARLYPDSKTFVDFQMRKDENATLSAFQELLDRTNHNPTKEDLQEFVVDFFDETSELEEWKPDDHKENPPFLAKIRDEGFREFAKALNDIWPTLARRVKPSVLEKPEQSSLVPMTHGFIVPGGRFKEIYYWDAYWIIEGLLITDMTETAKGMIENLIELLYKFGHIPNGSRWYYQERSQPPLLAAMIKLYYEKTKDIEFIRKYISALEKELEYWLDTHLIAFNKNDRVYTLLRYYIPSAGPRPESYYEDYELAQKLNKTTDPNDIYADLKSAAESGWDFSTRWFISESGDNSGNLTNLNTKNVIPVDLNAIFAGALQITANFQAILKNPRRAAHWGYMAEQWRSSIEQALWDEEDGVWHDYDILNNKPRRYFYTSNLAPLWMNAVEKPFLAKHGARVLEYLHESQALEYPGGVPVSLVNSGEQWDFPNAWPPEVSIVVTAIQNIGSEESSKLAKELAQVWVRACKSGFTEKKQMFEKYDALNAGKYGGGGEYTVQDGFGWSNGVVLEFLDRYGAVLTSVDSVDASANNGQSNEESETDSKEK
Summary
Catalytic Activity
alpha,alpha-trehalose + H2O = alpha-D-glucose + beta-D-glucose
Similarity
Belongs to the glycosyl hydrolase 37 family.
Keywords
Cell membrane
Complete proteome
Direct protein sequencing
Glycoprotein
Glycosidase
Hydrolase
Membrane
Reference proteome
Signal
Secreted
Feature
chain Trehalase
Uniprot
H9J822
P32358
A0A2D0WL58
A0A0L7LE05
C9S264
A0A1B2AQF5
+ More
A0A194QTL1 A0A194QAN3 A0A0U2KM65 A0A2A4JN84 A0A3Q8VR24 A0A0F6PB63 A3RLQ5 D2KHJ0 E9MYJ9 Q0ZIF5 E9MYK0 E9MYK2 E9MYJ8 A0A2H1WGC5 E9MYK1 B0M0J3 A0A212FIA5 L7WW66 A0A194QS77 A0A194QAV1 H9J823 R9YXT2 A0A2H1WC95 A0A076JZN4 D6WYI7 Q8MMG9 A0A2D0WLR3 P32359 A0A1W4XR71 A0A1Y1LGB5 A0A1D8I2M4 A0A2A4JMD9 A0A1B6DB06 A0A1B6E3A9 Q3MV18 A0A385MK80 A0A1B6FTE3 A0A2L1TG97 A0A2S2PXE7 B5ATV4 A0A2J7Q3Z7 A0A1B6FGD8 H9J4Z6 A0A2D0WLR5 A0A067QSZ4 A7XZC0 A0A1B6EY79 A0A1S6Q350 A0A2J7REE6 A0A1B6CFS1 D6WYI8 A0A1B6DGL1 E0W4A8 A0A2H8TFR3 J9JU09 A0A195FE62 A0A0K8SR01 A0A224X931 A0A195BF82 A0A0K8SQP5 G9I465 A0A1B6KBM1 A0A1B6KL82 T1HKP0 A0A0F6PAZ3 A0A0K8TDD3 A0A067QGU4 E2BGR0 A0A2W1BGC4 A0A146LQW6 A0A0A9WUU0 R9YY06 A0A2H8TKV2 A0A2J7PTL5 D2KHI9 A0A1D8I2M1 S4UQ18 A0A2H1WN76 A0A2J7PTK0 A0A1B6F1K5 X2G4I4 A0A1Y1LN31 A0A1Y1LKH8 C3UWP1 E2BSG4 M1RYJ3 A0A1B2AQF4 A0A212FIB4 A0A0C9S2J2 A0A3G2SGP7 J9JVF6 A0A2P8YXZ4 A0A084WJ24 A3RLQ4
A0A194QTL1 A0A194QAN3 A0A0U2KM65 A0A2A4JN84 A0A3Q8VR24 A0A0F6PB63 A3RLQ5 D2KHJ0 E9MYJ9 Q0ZIF5 E9MYK0 E9MYK2 E9MYJ8 A0A2H1WGC5 E9MYK1 B0M0J3 A0A212FIA5 L7WW66 A0A194QS77 A0A194QAV1 H9J823 R9YXT2 A0A2H1WC95 A0A076JZN4 D6WYI7 Q8MMG9 A0A2D0WLR3 P32359 A0A1W4XR71 A0A1Y1LGB5 A0A1D8I2M4 A0A2A4JMD9 A0A1B6DB06 A0A1B6E3A9 Q3MV18 A0A385MK80 A0A1B6FTE3 A0A2L1TG97 A0A2S2PXE7 B5ATV4 A0A2J7Q3Z7 A0A1B6FGD8 H9J4Z6 A0A2D0WLR5 A0A067QSZ4 A7XZC0 A0A1B6EY79 A0A1S6Q350 A0A2J7REE6 A0A1B6CFS1 D6WYI8 A0A1B6DGL1 E0W4A8 A0A2H8TFR3 J9JU09 A0A195FE62 A0A0K8SR01 A0A224X931 A0A195BF82 A0A0K8SQP5 G9I465 A0A1B6KBM1 A0A1B6KL82 T1HKP0 A0A0F6PAZ3 A0A0K8TDD3 A0A067QGU4 E2BGR0 A0A2W1BGC4 A0A146LQW6 A0A0A9WUU0 R9YY06 A0A2H8TKV2 A0A2J7PTL5 D2KHI9 A0A1D8I2M1 S4UQ18 A0A2H1WN76 A0A2J7PTK0 A0A1B6F1K5 X2G4I4 A0A1Y1LN31 A0A1Y1LKH8 C3UWP1 E2BSG4 M1RYJ3 A0A1B2AQF4 A0A212FIB4 A0A0C9S2J2 A0A3G2SGP7 J9JVF6 A0A2P8YXZ4 A0A084WJ24 A3RLQ4
EC Number
3.2.1.28
Pubmed
EMBL
BABH01020287
D13763
S73271
D86212
KU977455
ARD05073.1
+ More
JTDY01001509 KOB73697.1 CT955980 CBH09244.1 KU640203 ANY30160.1 KQ461155 KPJ08340.1 KQ459232 KPJ02593.1 KR149447 ALF03966.1 NWSH01000985 PCG73246.1 MH050739 AZM68711.1 KJ652557 AJK29979.1 EF426724 ABO20846.1 GU211891 ADA63846.1 HQ328815 ADT64093.1 DQ447188 ABE27189.1 HQ328816 ADT64094.1 HQ328818 ADT64096.1 HQ328814 ADT64092.1 ODYU01008486 SOQ52128.1 HQ328817 ADT64095.1 EU427311 ABY86218.1 AGBW02008409 OWR53472.1 KC469893 AGC92667.2 KPJ08342.1 KPJ02592.1 KC818237 AGO32658.1 ODYU01007692 SOQ50705.1 KJ789159 AII81931.1 KQ971362 EFA08991.1 AJ459958 KU977456 ARD05074.1 D11338 GEZM01058095 JAV71911.1 KU756285 AOT99588.1 PCG73245.1 GEDC01021380 GEDC01014488 JAS15918.1 JAS22810.1 GEDC01014640 GEDC01004890 JAS22658.1 JAS32408.1 AB162717 BAE45249.1 MG460307 AYA60780.1 GECZ01016299 JAS53470.1 KY697913 AVF19696.1 GGMS01000994 MBY70197.1 EU872435 ACF94698.1 NEVH01018420 PNF23300.1 GECZ01020512 JAS49257.1 BABH01025716 KU977457 ARD05075.1 KK852980 KDR13013.1 EU106080 ABU95354.1 GECZ01026821 JAS42948.1 KX371565 AQV08187.1 NEVH01005277 PNF39211.1 GEDC01025075 JAS12223.1 EFA08992.1 GEDC01012469 JAS24829.1 DS235886 EEB20464.1 GFXV01001141 MBW12946.1 ABLF02022564 KQ981673 KYN38309.1 GBRD01010280 JAG55544.1 GFTR01007580 JAW08846.1 KQ976500 KYM83241.1 GBRD01010278 GDHC01021605 GDHC01004455 JAG55546.1 JAP97023.1 JAQ14174.1 JN565077 AEW67359.1 GEBQ01031162 JAT08815.1 GEBQ01027776 JAT12201.1 ACPB03020626 KJ652558 AJK29980.1 GBRD01002317 JAG63504.1 KK853443 KDR07599.1 GL448204 EFN85130.1 KZ150067 PZC74122.1 GDHC01009483 JAQ09146.1 GBHO01031362 JAG12242.1 KC853750 AGO32660.1 GFXV01002874 MBW14679.1 NEVH01021237 PNF19664.1 GU211890 ADA63845.1 KU756284 AOT99587.1 JX312193 AGL34007.2 ODYU01009835 SOQ54523.1 PNF19666.1 GECZ01025694 JAS44075.1 KJ025078 AHN15422.1 GEZM01054932 GEZM01054931 GEZM01054929 GEZM01054927 JAV73335.1 GEZM01054933 GEZM01054930 GEZM01054928 GEZM01054926 JAV73338.1 FJ795020 ACP28173.1 GL450233 EFN81352.1 JX134487 AGG35943.1 KU640202 ANY30159.1 OWR53471.1 GBYB01015126 JAG84893.1 MG787168 AYO46921.1 ABLF02022430 PYGN01000293 PSN49112.1 ATLV01023963 ATLV01023964 ATLV01023965 KE525347 KFB50218.1 EF426723 ABO20845.1
JTDY01001509 KOB73697.1 CT955980 CBH09244.1 KU640203 ANY30160.1 KQ461155 KPJ08340.1 KQ459232 KPJ02593.1 KR149447 ALF03966.1 NWSH01000985 PCG73246.1 MH050739 AZM68711.1 KJ652557 AJK29979.1 EF426724 ABO20846.1 GU211891 ADA63846.1 HQ328815 ADT64093.1 DQ447188 ABE27189.1 HQ328816 ADT64094.1 HQ328818 ADT64096.1 HQ328814 ADT64092.1 ODYU01008486 SOQ52128.1 HQ328817 ADT64095.1 EU427311 ABY86218.1 AGBW02008409 OWR53472.1 KC469893 AGC92667.2 KPJ08342.1 KPJ02592.1 KC818237 AGO32658.1 ODYU01007692 SOQ50705.1 KJ789159 AII81931.1 KQ971362 EFA08991.1 AJ459958 KU977456 ARD05074.1 D11338 GEZM01058095 JAV71911.1 KU756285 AOT99588.1 PCG73245.1 GEDC01021380 GEDC01014488 JAS15918.1 JAS22810.1 GEDC01014640 GEDC01004890 JAS22658.1 JAS32408.1 AB162717 BAE45249.1 MG460307 AYA60780.1 GECZ01016299 JAS53470.1 KY697913 AVF19696.1 GGMS01000994 MBY70197.1 EU872435 ACF94698.1 NEVH01018420 PNF23300.1 GECZ01020512 JAS49257.1 BABH01025716 KU977457 ARD05075.1 KK852980 KDR13013.1 EU106080 ABU95354.1 GECZ01026821 JAS42948.1 KX371565 AQV08187.1 NEVH01005277 PNF39211.1 GEDC01025075 JAS12223.1 EFA08992.1 GEDC01012469 JAS24829.1 DS235886 EEB20464.1 GFXV01001141 MBW12946.1 ABLF02022564 KQ981673 KYN38309.1 GBRD01010280 JAG55544.1 GFTR01007580 JAW08846.1 KQ976500 KYM83241.1 GBRD01010278 GDHC01021605 GDHC01004455 JAG55546.1 JAP97023.1 JAQ14174.1 JN565077 AEW67359.1 GEBQ01031162 JAT08815.1 GEBQ01027776 JAT12201.1 ACPB03020626 KJ652558 AJK29980.1 GBRD01002317 JAG63504.1 KK853443 KDR07599.1 GL448204 EFN85130.1 KZ150067 PZC74122.1 GDHC01009483 JAQ09146.1 GBHO01031362 JAG12242.1 KC853750 AGO32660.1 GFXV01002874 MBW14679.1 NEVH01021237 PNF19664.1 GU211890 ADA63845.1 KU756284 AOT99587.1 JX312193 AGL34007.2 ODYU01009835 SOQ54523.1 PNF19666.1 GECZ01025694 JAS44075.1 KJ025078 AHN15422.1 GEZM01054932 GEZM01054931 GEZM01054929 GEZM01054927 JAV73335.1 GEZM01054933 GEZM01054930 GEZM01054928 GEZM01054926 JAV73338.1 FJ795020 ACP28173.1 GL450233 EFN81352.1 JX134487 AGG35943.1 KU640202 ANY30159.1 OWR53471.1 GBYB01015126 JAG84893.1 MG787168 AYO46921.1 ABLF02022430 PYGN01000293 PSN49112.1 ATLV01023963 ATLV01023964 ATLV01023965 KE525347 KFB50218.1 EF426723 ABO20845.1
Proteomes
Pfam
PF01204 Trehalase
Interpro
SUPFAM
SSF48208
SSF48208
Gene 3D
ProteinModelPortal
H9J822
P32358
A0A2D0WL58
A0A0L7LE05
C9S264
A0A1B2AQF5
+ More
A0A194QTL1 A0A194QAN3 A0A0U2KM65 A0A2A4JN84 A0A3Q8VR24 A0A0F6PB63 A3RLQ5 D2KHJ0 E9MYJ9 Q0ZIF5 E9MYK0 E9MYK2 E9MYJ8 A0A2H1WGC5 E9MYK1 B0M0J3 A0A212FIA5 L7WW66 A0A194QS77 A0A194QAV1 H9J823 R9YXT2 A0A2H1WC95 A0A076JZN4 D6WYI7 Q8MMG9 A0A2D0WLR3 P32359 A0A1W4XR71 A0A1Y1LGB5 A0A1D8I2M4 A0A2A4JMD9 A0A1B6DB06 A0A1B6E3A9 Q3MV18 A0A385MK80 A0A1B6FTE3 A0A2L1TG97 A0A2S2PXE7 B5ATV4 A0A2J7Q3Z7 A0A1B6FGD8 H9J4Z6 A0A2D0WLR5 A0A067QSZ4 A7XZC0 A0A1B6EY79 A0A1S6Q350 A0A2J7REE6 A0A1B6CFS1 D6WYI8 A0A1B6DGL1 E0W4A8 A0A2H8TFR3 J9JU09 A0A195FE62 A0A0K8SR01 A0A224X931 A0A195BF82 A0A0K8SQP5 G9I465 A0A1B6KBM1 A0A1B6KL82 T1HKP0 A0A0F6PAZ3 A0A0K8TDD3 A0A067QGU4 E2BGR0 A0A2W1BGC4 A0A146LQW6 A0A0A9WUU0 R9YY06 A0A2H8TKV2 A0A2J7PTL5 D2KHI9 A0A1D8I2M1 S4UQ18 A0A2H1WN76 A0A2J7PTK0 A0A1B6F1K5 X2G4I4 A0A1Y1LN31 A0A1Y1LKH8 C3UWP1 E2BSG4 M1RYJ3 A0A1B2AQF4 A0A212FIB4 A0A0C9S2J2 A0A3G2SGP7 J9JVF6 A0A2P8YXZ4 A0A084WJ24 A3RLQ4
A0A194QTL1 A0A194QAN3 A0A0U2KM65 A0A2A4JN84 A0A3Q8VR24 A0A0F6PB63 A3RLQ5 D2KHJ0 E9MYJ9 Q0ZIF5 E9MYK0 E9MYK2 E9MYJ8 A0A2H1WGC5 E9MYK1 B0M0J3 A0A212FIA5 L7WW66 A0A194QS77 A0A194QAV1 H9J823 R9YXT2 A0A2H1WC95 A0A076JZN4 D6WYI7 Q8MMG9 A0A2D0WLR3 P32359 A0A1W4XR71 A0A1Y1LGB5 A0A1D8I2M4 A0A2A4JMD9 A0A1B6DB06 A0A1B6E3A9 Q3MV18 A0A385MK80 A0A1B6FTE3 A0A2L1TG97 A0A2S2PXE7 B5ATV4 A0A2J7Q3Z7 A0A1B6FGD8 H9J4Z6 A0A2D0WLR5 A0A067QSZ4 A7XZC0 A0A1B6EY79 A0A1S6Q350 A0A2J7REE6 A0A1B6CFS1 D6WYI8 A0A1B6DGL1 E0W4A8 A0A2H8TFR3 J9JU09 A0A195FE62 A0A0K8SR01 A0A224X931 A0A195BF82 A0A0K8SQP5 G9I465 A0A1B6KBM1 A0A1B6KL82 T1HKP0 A0A0F6PAZ3 A0A0K8TDD3 A0A067QGU4 E2BGR0 A0A2W1BGC4 A0A146LQW6 A0A0A9WUU0 R9YY06 A0A2H8TKV2 A0A2J7PTL5 D2KHI9 A0A1D8I2M1 S4UQ18 A0A2H1WN76 A0A2J7PTK0 A0A1B6F1K5 X2G4I4 A0A1Y1LN31 A0A1Y1LKH8 C3UWP1 E2BSG4 M1RYJ3 A0A1B2AQF4 A0A212FIB4 A0A0C9S2J2 A0A3G2SGP7 J9JVF6 A0A2P8YXZ4 A0A084WJ24 A3RLQ4
PDB
5Z6H
E-value=4.87414e-75,
Score=717
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Basolateral cell membrane
Secreted
Secreted
SignalP
Position: 1 - 15,
Likelihood: 0.998917
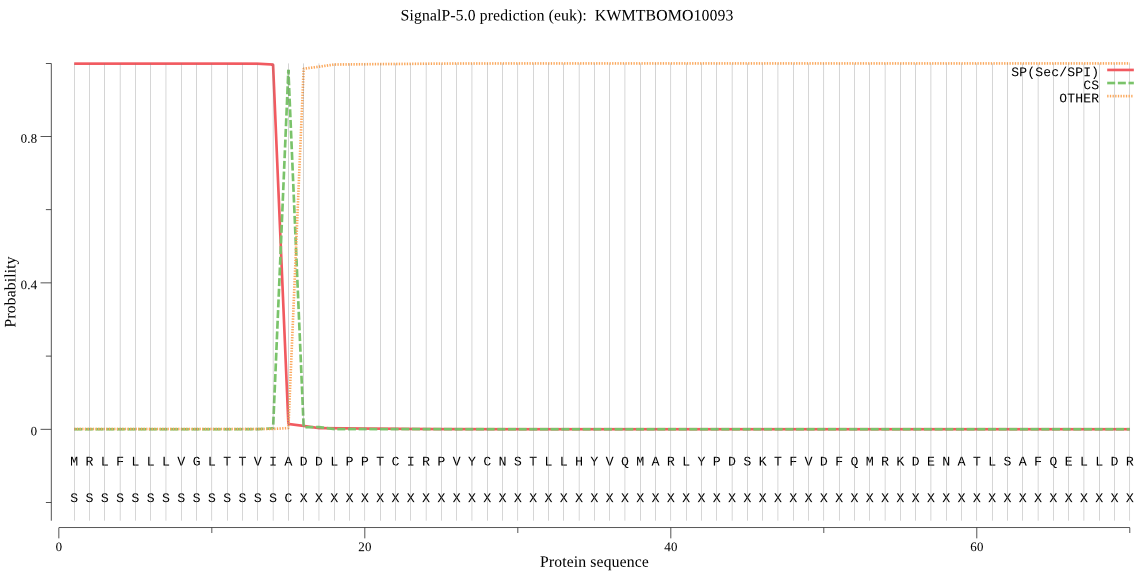
Length:
579
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02588
Exp number, first 60 AAs:
0.0068
Total prob of N-in:
0.00038
outside
1 - 579
Population Genetic Test Statistics
Pi
164.098744
Theta
163.345709
Tajima's D
0.685049
CLR
0.348433
CSRT
0.565121743912804
Interpretation
Uncertain
