Gene
KWMTBOMO10092
Pre Gene Modal
BGIBMGA005665
Annotation
Trehalase_[Operophtera_brumata]
Full name
Trehalase
Alternative Name
Alpha-trehalose glucohydrolase
Alpha,alpha-trehalase
Alpha,alpha-trehalose glucohydrolase
Alpha,alpha-trehalase
Alpha,alpha-trehalose glucohydrolase
Location in the cell
Cytoplasmic Reliability : 1.417
Sequence
CDS
ATGTATAAATCACGTTCTATACGCCTGTACCGCGTGATCGGGCTGCTTGGAGCATGCGCTCAAGCTGTGCACATTGTTCCCGCTTGTAACTCCTCAATACATTGCTCCGGTGAACTCCTGCATAAAGTTCAGCTGGCCCGAATCTTTCCCGACTCGAAGACTTTTGTCGACTTGAAACTAAAGTATCCGGAATATCAAACTCTCGCAGATTTCGCTGAATTGATGAAAGAGACACATCATGAACCCAGTCGCGCTAGTCTAGCTCGATTTGTGGACAGTCATTTTATGGAAGGGAATGAACTCGAAGATTGGGATCCACCTGATTTTGATCCCGAGCCGCCAATCCTGGAAAACATCACAGATGTCAGACTCAAGCGATTCGCTAAAGGCATTATTAGCATTTGGGCCAAATTAGGCCGAAAAGTTAGGCCAGATCTCATTCAAGAAGAAGACCAGTATAGTTTCATTTCGGTGCCCAACGGTTTTATTGTTCCTGGTGGAAGATTCAAAGAGCTATATTACTGGGATTCGTTTTGGATGATACGTGGATTAATTATTAGTAACATGATGCAGACAGCAAAAGGGATGATAGAAAATTTGTTACATTTGGTGGAAAAAGTTGGCTATATACCTAATGGCAGCCGAATTTATTACCTAGGCCGCAGCCAACCCCCCCTACTCACAGCAATGGTCGCAAGTTATTTTGCAGCCACCGGCGACATCGTGTGGCTTGAACGACACATCAGCACCGTCGAAAAAGAACTTCAATATTGGCTCGACAAAAAGAAAGTCACTGTTACAGTTGAAGGTAAAAAATATATTCTTCTCCGCTATCTTTCTGACAGAAGAAGTAAAGGACCTCGTCCAGAATCTTATTATGAAGACTATACTAACGCACAAGTGTTACCCACTGAAGAGTTGCGAGAAGATTTCTACAATGAAATGAAAAGTGCAGCAGAAAGTGGATGGGACTTTTCGACTCGATGGTTTGTTACTGCCAAGAATGAAACTGTTGGCTGCTTGACTGACGTTCACGCGACTCGTATCGTGCCTGTCGATCTAAATGCTATCTTCGCTGGGGCCCTTGAACTTGCCGGTGACTTCAGGTATCATCTAAAAGACAGGCGTGAAGCGAAAAAATGGTGGAGTCTTGCCAAATATTGGAGGAACGCGATTGACGCCGTGCTTTGGGATGCCAATGATGGTGTTTGGTACGATTACGACACTCAAGCTAAAGCTTCGAGACGTCACTTTTATCCAAGCTCCGCGACTCCGTTATGGTCAGGAGCCGTGGAAAGCTCCGAAGCTCCACGGTATGCGGCGAAATTCGTAAAATATCTCCTGTCATCTGGCGCGTTAAGCTTCCCTGGTGGGATCCCAGCTTCCGTGCTACATTCAGGTGAACAGTGGGACTATCCAAATGCTTGGCCTCCTTTACAGAGCATCTTGATCGGTGGCTTAGAGATGAGCGGAAATGAAGAGGCAAGGAGGTTGGCGAGAGAACAAGCAAGAATATGGATTCGCGCCAATTATATCGGTTTTACTACATGGAATAAGATGTTTGAGAAGTACAGTGTAGTAGAACCAGGGCATCACGGCAGCGGAGGGGAGTATATAGTGCAGGATGGTTTCGGGTGGACCAATGGGGTCGCCCTAGAACTCTTGCAAATGTACGGGAAGGAGATGACGTTGGATGACAGCCCCGAAAATTTTCCGTACCTAACGGTGGTAAAATAA
Protein
MYKSRSIRLYRVIGLLGACAQAVHIVPACNSSIHCSGELLHKVQLARIFPDSKTFVDLKLKYPEYQTLADFAELMKETHHEPSRASLARFVDSHFMEGNELEDWDPPDFDPEPPILENITDVRLKRFAKGIISIWAKLGRKVRPDLIQEEDQYSFISVPNGFIVPGGRFKELYYWDSFWMIRGLIISNMMQTAKGMIENLLHLVEKVGYIPNGSRIYYLGRSQPPLLTAMVASYFAATGDIVWLERHISTVEKELQYWLDKKKVTVTVEGKKYILLRYLSDRRSKGPRPESYYEDYTNAQVLPTEELREDFYNEMKSAAESGWDFSTRWFVTAKNETVGCLTDVHATRIVPVDLNAIFAGALELAGDFRYHLKDRREAKKWWSLAKYWRNAIDAVLWDANDGVWYDYDTQAKASRRHFYPSSATPLWSGAVESSEAPRYAAKFVKYLLSSGALSFPGGIPASVLHSGEQWDYPNAWPPLQSILIGGLEMSGNEEARRLAREQARIWIRANYIGFTTWNKMFEKYSVVEPGHHGSGGEYIVQDGFGWTNGVALELLQMYGKEMTLDDSPENFPYLTVVK
Summary
Catalytic Activity
alpha,alpha-trehalose + H2O = alpha-D-glucose + beta-D-glucose
Similarity
Belongs to the glycosyl hydrolase 37 family.
Keywords
Cell membrane
Complete proteome
Direct protein sequencing
Glycoprotein
Glycosidase
Hydrolase
Membrane
Reference proteome
Signal
Secreted
Feature
chain Trehalase
Uniprot
H9J823
A0A0L7LE05
A0A194QS77
A0A194QAV1
L7WW66
A0A2D0WLR3
+ More
A0A2H1WC95 A0A2A4JMD9 A0A212FIB4 A0A194QAN3 A0A194QTL1 Q0ZIF5 D2KHJ0 C9S264 P32358 H9J822 A0A3Q8VR24 A0A1B2AQF5 A0A2H1WGC5 A0A2D0WL58 B0M0J3 A0A0U2KM65 A0A2A4JN84 E9MYJ9 A0A0F6PB63 A3RLQ5 D6WYI7 E9MYK2 E9MYK0 P32359 E9MYK1 R9YXT2 E9MYJ8 A0A1Y1LGB5 E0W4A8 A0A1W4XR71 A0A212FIA5 A0A2H8TFR3 J9JU09 A0A067QSZ4 G9I465 A0A1B6KBM1 A0A1B6KL82 A0A288VJ26 A0A288VJ85 X2G4I4 D6WYI8 S4UQ18 A0A2S2PXE7 A0A067QGU4 A0A2J7Q3Z7 A0A1D8I2M1 A0A1S6Q350 A0A2J7REE6 A0A1B6F1K5 A0A1S3D5M3 A0A1S3D5V1 E2BSG4 A0A1D8I2M4 A0A1S3D5L1 A0A1X9IF78 A0A1S3D5U6 A0A0N7CXG3 T1HKP0 A0A195FE62 A0A146LQW6 A0A385MK80 A0A0A9WUU0 A0A0K8TDD3 A0A2L1TG97 A0A2A3ECD2 J3JYK0 A0A076JZN4 A0A2R7X5I1 A0A1B6DUA6 A0A1B6EEF7 K7R0G4 A0A288VJ48 Q8MMG9 A0A154P327 I3WEV3 V5GU59 I7CKW6 E9I9Z8 A0A2K8JSN4 A0A195BF82 R9YY06 U4TSF0 A0A0L7QQW6 A0A2P8YXZ4 A0A1B6E3A9 A0A1B6DB06 N6TW80 A0A1W4XME4 A0A023F5Y3 A0A1B6EC02 A0A1W4XCC1 A0A1B6FTE3
A0A2H1WC95 A0A2A4JMD9 A0A212FIB4 A0A194QAN3 A0A194QTL1 Q0ZIF5 D2KHJ0 C9S264 P32358 H9J822 A0A3Q8VR24 A0A1B2AQF5 A0A2H1WGC5 A0A2D0WL58 B0M0J3 A0A0U2KM65 A0A2A4JN84 E9MYJ9 A0A0F6PB63 A3RLQ5 D6WYI7 E9MYK2 E9MYK0 P32359 E9MYK1 R9YXT2 E9MYJ8 A0A1Y1LGB5 E0W4A8 A0A1W4XR71 A0A212FIA5 A0A2H8TFR3 J9JU09 A0A067QSZ4 G9I465 A0A1B6KBM1 A0A1B6KL82 A0A288VJ26 A0A288VJ85 X2G4I4 D6WYI8 S4UQ18 A0A2S2PXE7 A0A067QGU4 A0A2J7Q3Z7 A0A1D8I2M1 A0A1S6Q350 A0A2J7REE6 A0A1B6F1K5 A0A1S3D5M3 A0A1S3D5V1 E2BSG4 A0A1D8I2M4 A0A1S3D5L1 A0A1X9IF78 A0A1S3D5U6 A0A0N7CXG3 T1HKP0 A0A195FE62 A0A146LQW6 A0A385MK80 A0A0A9WUU0 A0A0K8TDD3 A0A2L1TG97 A0A2A3ECD2 J3JYK0 A0A076JZN4 A0A2R7X5I1 A0A1B6DUA6 A0A1B6EEF7 K7R0G4 A0A288VJ48 Q8MMG9 A0A154P327 I3WEV3 V5GU59 I7CKW6 E9I9Z8 A0A2K8JSN4 A0A195BF82 R9YY06 U4TSF0 A0A0L7QQW6 A0A2P8YXZ4 A0A1B6E3A9 A0A1B6DB06 N6TW80 A0A1W4XME4 A0A023F5Y3 A0A1B6EC02 A0A1W4XCC1 A0A1B6FTE3
EC Number
3.2.1.28
Pubmed
EMBL
BABH01020287
JTDY01001509
KOB73697.1
KQ461155
KPJ08342.1
KQ459232
+ More
KPJ02592.1 KC469893 AGC92667.2 KU977456 ARD05074.1 ODYU01007692 SOQ50705.1 NWSH01000985 PCG73245.1 AGBW02008409 OWR53471.1 KPJ02593.1 KPJ08340.1 DQ447188 ABE27189.1 GU211891 ADA63846.1 CT955980 CBH09244.1 D13763 S73271 D86212 MH050739 AZM68711.1 KU640203 ANY30160.1 ODYU01008486 SOQ52128.1 KU977455 ARD05073.1 EU427311 ABY86218.1 KR149447 ALF03966.1 PCG73246.1 HQ328815 ADT64093.1 KJ652557 AJK29979.1 EF426724 ABO20846.1 KQ971362 EFA08991.1 HQ328818 ADT64096.1 HQ328816 ADT64094.1 D11338 HQ328817 ADT64095.1 KC818237 AGO32658.1 HQ328814 ADT64092.1 GEZM01058095 JAV71911.1 DS235886 EEB20464.1 OWR53472.1 GFXV01001141 MBW12946.1 ABLF02022564 KK852980 KDR13013.1 JN565077 AEW67359.1 GEBQ01031162 JAT08815.1 GEBQ01027776 JAT12201.1 KY400002 AQY15494.1 KY400001 KY400003 AQY15493.1 KJ025078 AHN15422.1 EFA08992.1 JX312193 AGL34007.2 GGMS01000994 MBY70197.1 KK853443 KDR07599.1 NEVH01018420 PNF23300.1 KU756284 AOT99587.1 KX371565 AQV08187.1 NEVH01005277 PNF39211.1 GECZ01025694 JAS44075.1 GL450233 EFN81352.1 KU756285 AOT99588.1 KX349225 AOT82130.1 KP318742 AKQ19374.1 ACPB03020626 KQ981673 KYN38309.1 GDHC01009483 JAQ09146.1 MG460307 AYA60780.1 GBHO01031362 JAG12242.1 GBRD01002317 JAG63504.1 KY697913 AVF19696.1 KZ288288 PBC29350.1 APGK01035671 APGK01035672 APGK01035673 BT128329 KB740928 AEE63287.1 ENN77913.1 KJ789159 AII81931.1 KK857312 PTY27036.1 GEDC01008038 JAS29260.1 GEDC01000986 JAS36312.1 JX024262 AFV79627.1 KY400004 KY400005 KY400006 AQY15496.1 AJ459958 KQ434796 KZC05540.1 JQ027051 AFL03410.1 GALX01000767 JAB67699.1 JX204291 AFO54713.1 GL761864 EFZ22607.1 KY031201 ATU82952.1 KQ976500 KYM83241.1 KC853750 AGO32660.1 KB631579 ERL84424.1 KQ414784 KOC61023.1 PYGN01000293 PSN49112.1 GEDC01014640 GEDC01004890 JAS22658.1 JAS32408.1 GEDC01021380 GEDC01014488 JAS15918.1 JAS22810.1 APGK01058678 KB741291 ENN70552.1 GBBI01002113 JAC16599.1 GEDC01001832 JAS35466.1 GECZ01016299 JAS53470.1
KPJ02592.1 KC469893 AGC92667.2 KU977456 ARD05074.1 ODYU01007692 SOQ50705.1 NWSH01000985 PCG73245.1 AGBW02008409 OWR53471.1 KPJ02593.1 KPJ08340.1 DQ447188 ABE27189.1 GU211891 ADA63846.1 CT955980 CBH09244.1 D13763 S73271 D86212 MH050739 AZM68711.1 KU640203 ANY30160.1 ODYU01008486 SOQ52128.1 KU977455 ARD05073.1 EU427311 ABY86218.1 KR149447 ALF03966.1 PCG73246.1 HQ328815 ADT64093.1 KJ652557 AJK29979.1 EF426724 ABO20846.1 KQ971362 EFA08991.1 HQ328818 ADT64096.1 HQ328816 ADT64094.1 D11338 HQ328817 ADT64095.1 KC818237 AGO32658.1 HQ328814 ADT64092.1 GEZM01058095 JAV71911.1 DS235886 EEB20464.1 OWR53472.1 GFXV01001141 MBW12946.1 ABLF02022564 KK852980 KDR13013.1 JN565077 AEW67359.1 GEBQ01031162 JAT08815.1 GEBQ01027776 JAT12201.1 KY400002 AQY15494.1 KY400001 KY400003 AQY15493.1 KJ025078 AHN15422.1 EFA08992.1 JX312193 AGL34007.2 GGMS01000994 MBY70197.1 KK853443 KDR07599.1 NEVH01018420 PNF23300.1 KU756284 AOT99587.1 KX371565 AQV08187.1 NEVH01005277 PNF39211.1 GECZ01025694 JAS44075.1 GL450233 EFN81352.1 KU756285 AOT99588.1 KX349225 AOT82130.1 KP318742 AKQ19374.1 ACPB03020626 KQ981673 KYN38309.1 GDHC01009483 JAQ09146.1 MG460307 AYA60780.1 GBHO01031362 JAG12242.1 GBRD01002317 JAG63504.1 KY697913 AVF19696.1 KZ288288 PBC29350.1 APGK01035671 APGK01035672 APGK01035673 BT128329 KB740928 AEE63287.1 ENN77913.1 KJ789159 AII81931.1 KK857312 PTY27036.1 GEDC01008038 JAS29260.1 GEDC01000986 JAS36312.1 JX024262 AFV79627.1 KY400004 KY400005 KY400006 AQY15496.1 AJ459958 KQ434796 KZC05540.1 JQ027051 AFL03410.1 GALX01000767 JAB67699.1 JX204291 AFO54713.1 GL761864 EFZ22607.1 KY031201 ATU82952.1 KQ976500 KYM83241.1 KC853750 AGO32660.1 KB631579 ERL84424.1 KQ414784 KOC61023.1 PYGN01000293 PSN49112.1 GEDC01014640 GEDC01004890 JAS22658.1 JAS32408.1 GEDC01021380 GEDC01014488 JAS15918.1 JAS22810.1 APGK01058678 KB741291 ENN70552.1 GBBI01002113 JAC16599.1 GEDC01001832 JAS35466.1 GECZ01016299 JAS53470.1
Proteomes
Pfam
PF01204 Trehalase
Interpro
SUPFAM
SSF48208
SSF48208
Gene 3D
ProteinModelPortal
H9J823
A0A0L7LE05
A0A194QS77
A0A194QAV1
L7WW66
A0A2D0WLR3
+ More
A0A2H1WC95 A0A2A4JMD9 A0A212FIB4 A0A194QAN3 A0A194QTL1 Q0ZIF5 D2KHJ0 C9S264 P32358 H9J822 A0A3Q8VR24 A0A1B2AQF5 A0A2H1WGC5 A0A2D0WL58 B0M0J3 A0A0U2KM65 A0A2A4JN84 E9MYJ9 A0A0F6PB63 A3RLQ5 D6WYI7 E9MYK2 E9MYK0 P32359 E9MYK1 R9YXT2 E9MYJ8 A0A1Y1LGB5 E0W4A8 A0A1W4XR71 A0A212FIA5 A0A2H8TFR3 J9JU09 A0A067QSZ4 G9I465 A0A1B6KBM1 A0A1B6KL82 A0A288VJ26 A0A288VJ85 X2G4I4 D6WYI8 S4UQ18 A0A2S2PXE7 A0A067QGU4 A0A2J7Q3Z7 A0A1D8I2M1 A0A1S6Q350 A0A2J7REE6 A0A1B6F1K5 A0A1S3D5M3 A0A1S3D5V1 E2BSG4 A0A1D8I2M4 A0A1S3D5L1 A0A1X9IF78 A0A1S3D5U6 A0A0N7CXG3 T1HKP0 A0A195FE62 A0A146LQW6 A0A385MK80 A0A0A9WUU0 A0A0K8TDD3 A0A2L1TG97 A0A2A3ECD2 J3JYK0 A0A076JZN4 A0A2R7X5I1 A0A1B6DUA6 A0A1B6EEF7 K7R0G4 A0A288VJ48 Q8MMG9 A0A154P327 I3WEV3 V5GU59 I7CKW6 E9I9Z8 A0A2K8JSN4 A0A195BF82 R9YY06 U4TSF0 A0A0L7QQW6 A0A2P8YXZ4 A0A1B6E3A9 A0A1B6DB06 N6TW80 A0A1W4XME4 A0A023F5Y3 A0A1B6EC02 A0A1W4XCC1 A0A1B6FTE3
A0A2H1WC95 A0A2A4JMD9 A0A212FIB4 A0A194QAN3 A0A194QTL1 Q0ZIF5 D2KHJ0 C9S264 P32358 H9J822 A0A3Q8VR24 A0A1B2AQF5 A0A2H1WGC5 A0A2D0WL58 B0M0J3 A0A0U2KM65 A0A2A4JN84 E9MYJ9 A0A0F6PB63 A3RLQ5 D6WYI7 E9MYK2 E9MYK0 P32359 E9MYK1 R9YXT2 E9MYJ8 A0A1Y1LGB5 E0W4A8 A0A1W4XR71 A0A212FIA5 A0A2H8TFR3 J9JU09 A0A067QSZ4 G9I465 A0A1B6KBM1 A0A1B6KL82 A0A288VJ26 A0A288VJ85 X2G4I4 D6WYI8 S4UQ18 A0A2S2PXE7 A0A067QGU4 A0A2J7Q3Z7 A0A1D8I2M1 A0A1S6Q350 A0A2J7REE6 A0A1B6F1K5 A0A1S3D5M3 A0A1S3D5V1 E2BSG4 A0A1D8I2M4 A0A1S3D5L1 A0A1X9IF78 A0A1S3D5U6 A0A0N7CXG3 T1HKP0 A0A195FE62 A0A146LQW6 A0A385MK80 A0A0A9WUU0 A0A0K8TDD3 A0A2L1TG97 A0A2A3ECD2 J3JYK0 A0A076JZN4 A0A2R7X5I1 A0A1B6DUA6 A0A1B6EEF7 K7R0G4 A0A288VJ48 Q8MMG9 A0A154P327 I3WEV3 V5GU59 I7CKW6 E9I9Z8 A0A2K8JSN4 A0A195BF82 R9YY06 U4TSF0 A0A0L7QQW6 A0A2P8YXZ4 A0A1B6E3A9 A0A1B6DB06 N6TW80 A0A1W4XME4 A0A023F5Y3 A0A1B6EC02 A0A1W4XCC1 A0A1B6FTE3
PDB
5Z6H
E-value=1.49121e-66,
Score=644
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Basolateral cell membrane
Secreted
Secreted
SignalP
Position: 1 - 22,
Likelihood: 0.796703
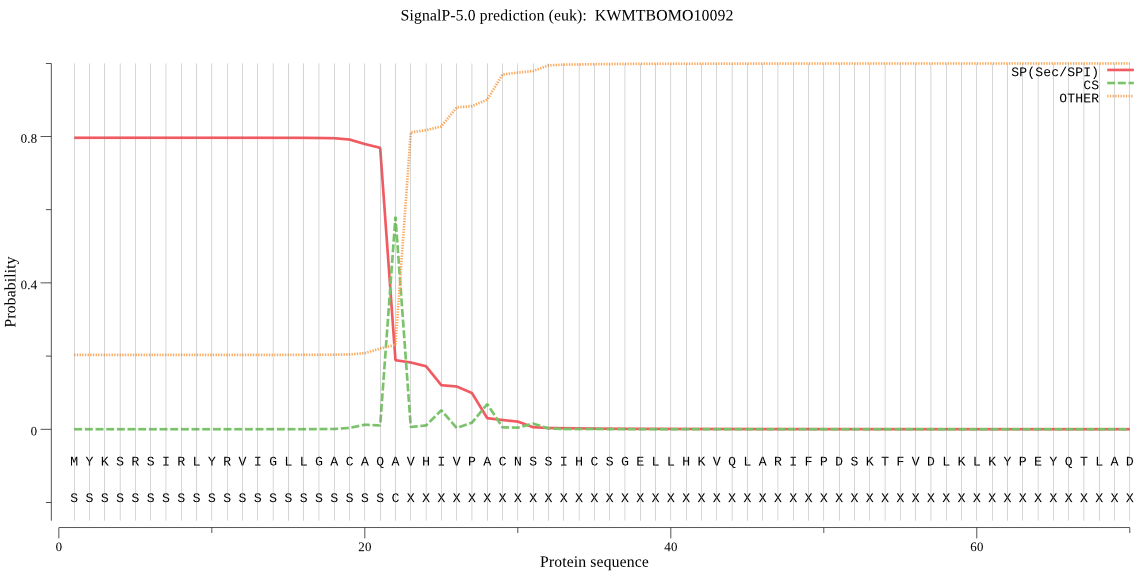
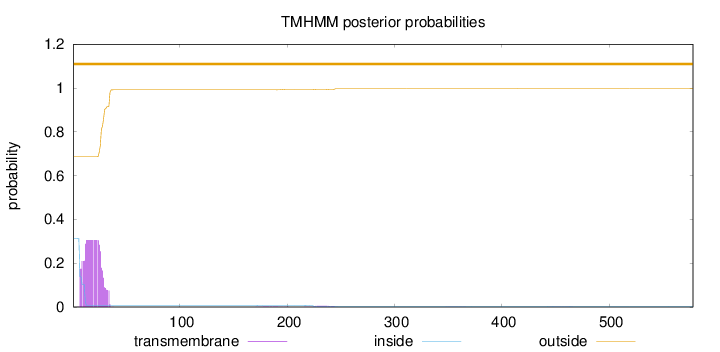
Length:
578
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.51790999999997
Exp number, first 60 AAs:
6.36627
Total prob of N-in:
0.31330
outside
1 - 578
Population Genetic Test Statistics
Pi
172.948809
Theta
15.674553
Tajima's D
0.717947
CLR
0.247659
CSRT
0.587670616469177
Interpretation
Uncertain
