Gene
KWMTBOMO10091
Pre Gene Modal
BGIBMGA005666
Annotation
PREDICTED:_ATP-dependent_DNA_helicase_Q1-like_[Amyelois_transitella]
Full name
ATP-dependent DNA helicase
+ More
ATP-dependent DNA helicase Q1
Glycogen [starch] synthase
ATP-dependent DNA helicase Q1
Glycogen [starch] synthase
Alternative Name
DNA-dependent ATPase Q1
RecQ protein-like 1
RecQ protein-like 1
Location in the cell
Nuclear Reliability : 2.448
Sequence
CDS
ATGAAGACCATAGAAGAACTACAAAATGAACTTATCAGCGTTAACGACGAATTGCAAAAGGTCGATGCTGAACTTGAGAAATGGAGGACCCGGCAAAGAAAATTGCACGAAAAACGCAATAGTTTAAAAAATACTATTAATACTATAAAATCTGAAACATTAGCGAGCGTTGACTGGGCTGGTACTGAATACGAGTGGTCGGAAGACGTCAAAAATGTTTTAAAAGAAAAATTTCGATTAAAAACTTTTCGTGCGAAACAAATGAGCGCGATTAATTCCACGTTATCAGGCCAACACGCGATAGTCGTGATGCCGACAGGCGCCGGTAAGAGTCTTTGCTACCAGCTACCTGCACTCGTTAGGCCTGGACTAACAGTGGTCGTGTCACCACTCGTGTCGCTTATGGAAGACCAAGTAAGATCTCTCACAGATAAGAAAATAAGCGCCAAATTGATAACCAGCACTAGTCCAAAAGAGGAAACGTCTGCTGTACTGAACATGCTAAAAGACAAATCTGCCACTGTAAAACTATTATATGTAACTCCGGAACGTTTTGCTAAAAGTAAGAGATTCATGTCGAATTTGCAAAAATGTTATACTGATGGAAGCCTGCAAAGAATTGCTATTGATGAGGTTCACTGTTGTTCCCAGTGGGGTCATGACTTTAGACCTGATTATAAGTATCTTGGGATACTGACTCAAATATTTCCCAACATTCCAATATTGGGGTTAACTGCGACAGCTACTGCTCATGTTCTAAATGATGTTCAAAAAATACTAAACATACCTGGTTGCTTAATCATAAAATCTACATTTAATCGTCCTAATTTGTATTACAAAATTCTAGAGAAACCAACATCCCAAGATGATTGTCTCATTATATTAGAAAAACTTTTGAAATACAGGTACAAAGATCAATCTGGAATAATCTACACAAATAGCATAAAAGATGCTGAGGAAATAGCTGAAGGACTAAGGAAAAAAGGTCTAAAAGTGGCTTGTTATCATGCTAATTTAGATGCAGATATTAGATCAAAGGTCCATATCCGATGGCATGAACAACATTACCAAGCCATAGTTGCAACTGTAGCATTTGGCATGGGAATTGATAAACCAGATGTCAGGTTTGTCATACACCACACTATCAGCAAATCAATGGAAAACTACTACCAAGAGAGTGGAAGAGCTGGACGAGATGGTCTCAGAGCTGAGTGCATAACCCTGTACAGAATGCAGGACATATTTAAAGTGAGCACTATGGTATTTCAGTCTTCAGCTGGTAGCACTGATCATTTATATGAAATGGTGAAATATTGTCTTAATGGAACATTATGTCGGAGGCAAATTATTGCAAAACATTTTGATGAAGATTGGGGTGATAGTGACTGCAATAAGATGTGTGACGTTTGCAGTACCAGTCAATCAGATTCACCCAAAGATTTGGATCTTAGAGCTCATTGTAGGATTATTGCTCATATTCTAGAAAATGCTGAGAAGCTAGACACCAAATTAACAGCTCAGAAATTACTTGATGCATGGTTTTTAAAAGGGCCAGTAAATTTAAGACATAAAGGAAAGGAGCCAAATTTTCCTCGTGTTTTAGGTGAAGATGTTATAGCATTTTTATTAATAAACGGTTATTTAGTAGAAGATTTTCATTTCACTGCTTATTCGACTATAAGCTACCTAAAAATAGGTCCTGAAATGTCAGGAGTAAATAATGATGGCTTTAAGCTTAAAATGCGAGTTAGAGTTTATTCCCATTTTGAATTGAGCAAGCTGGCACCTACTGAATTGTTTGAAGAAACAGATAATAAATCAGTGAAAAGGAAAACAGAACCACCACCGCAAGTAAAGGTTAAGAGAAGATTGGTTGAAATTGATGATTAA
Protein
MKTIEELQNELISVNDELQKVDAELEKWRTRQRKLHEKRNSLKNTINTIKSETLASVDWAGTEYEWSEDVKNVLKEKFRLKTFRAKQMSAINSTLSGQHAIVVMPTGAGKSLCYQLPALVRPGLTVVVSPLVSLMEDQVRSLTDKKISAKLITSTSPKEETSAVLNMLKDKSATVKLLYVTPERFAKSKRFMSNLQKCYTDGSLQRIAIDEVHCCSQWGHDFRPDYKYLGILTQIFPNIPILGLTATATAHVLNDVQKILNIPGCLIIKSTFNRPNLYYKILEKPTSQDDCLIILEKLLKYRYKDQSGIIYTNSIKDAEEIAEGLRKKGLKVACYHANLDADIRSKVHIRWHEQHYQAIVATVAFGMGIDKPDVRFVIHHTISKSMENYYQESGRAGRDGLRAECITLYRMQDIFKVSTMVFQSSAGSTDHLYEMVKYCLNGTLCRRQIIAKHFDEDWGDSDCNKMCDVCSTSQSDSPKDLDLRAHCRIIAHILENAEKLDTKLTAQKLLDAWFLKGPVNLRHKGKEPNFPRVLGEDVIAFLLINGYLVEDFHFTAYSTISYLKIGPEMSGVNNDGFKLKMRVRVYSHFELSKLAPTELFEETDNKSVKRKTEPPPQVKVKRRLVEIDD
Summary
Description
DNA helicase that may play a role in the repair of DNA that is damaged by ultraviolet light or other mutagens. Exhibits a magnesium-dependent ATP-dependent DNA-helicase activity that unwinds single- and double-stranded DNA in a 3'-5' direction (By similarity).
Transfers the glycosyl residue from UDP-Glc to the non-reducing end of alpha-1,4-glucan.
Transfers the glycosyl residue from UDP-Glc to the non-reducing end of alpha-1,4-glucan.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
[(1->4)-alpha-D-glucosyl](n) + UDP-alpha-D-glucose = [(1->4)-alpha-D-glucosyl](n+1) + H(+) + UDP
[(1->4)-alpha-D-glucosyl](n) + UDP-alpha-D-glucose = [(1->4)-alpha-D-glucosyl](n+1) + H(+) + UDP
Subunit
Interacts with EXO1 and MLH1.
Similarity
Belongs to the helicase family. RecQ subfamily.
Belongs to the glycosyltransferase 3 family.
Belongs to the glycosyltransferase 3 family.
Keywords
Acetylation
ATP-binding
Complete proteome
DNA-binding
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Feature
chain ATP-dependent DNA helicase
Uniprot
H9J824
A0A212FIK8
A0A2A4JJE6
A0A0L7LGS0
A0A194QSN4
A0A2H1WI67
+ More
L7X1M6 C9S278 A0A194QAP1 E3UKN2 A0A2J7Q8Y3 A0A1B6JNR2 A0A1B6CUY3 A0A2P8YQL9 A0A1B6M5M1 A0A3L8DC02 A0A026WLE0 A0A067RX24 E0VZQ3 A0A2A3EM88 A0A087ZZI7 F4WEW0 A0A151X9X4 E9JCN0 A0A151JNL4 A0A151K1E3 W5JVH8 A0A310SCJ2 A0A154PK42 A0A0J7KYL3 A0A232FL31 A0A0L7QNC7 A0A336MJW4 A0A336L3W0 K7J5G3 A0A195B5U2 A0A158NWX1 A0A0P5DHV1 A0A182FUS3 E9G2Y5 A0A0P4VRU5 A0A0P5HZH7 A0A1Q3F0B8 A0A2J7Q8Y0 A0A0P5C8B0 B0XD44 A0A291S6Y1 A0A0P5B466 A0A087SWY7 A0A1B0CJD7 A0A1J1J5C0 A0A0C9RZJ3 A0JN36 J3S495 A0A2R5LAI8 A0A0P5ZDE8 K7F4Y8 H0VZ66 A0A0P5Z9P4 K7F4Y6 A0A1B0DNX2 Q6AYJ1 G1NLK6 A0A2Y9NEU0 A0A250YKF7 A0A341ADV5 A0A2U4BG13 T1E5X8 G1P5G3 M3XM99 A0A091ELU9 A0A2Y9FPS5 M3WQ95 A0A0B8RW48 A0A287BGJ9 A0A384B6I2 Q8AYS4 F1PNP1 A0A3Q7T7K4 A0A091X1R1 L5KKB3 A0A084VID9 A0A340WP44 A0A3Q7WV68 A0A384C948 A0A2D4PLK1 W5QGJ8 A0A091RGT0 A0A226N6V9 A0A182PC96 A0A1S3FXI9 L8INL4 A0A210PLF8 R0KZK1 A0A2S2QP09 G5BEF7 G1MCS7 F1SR01 S7N182 A0A0A0A487
L7X1M6 C9S278 A0A194QAP1 E3UKN2 A0A2J7Q8Y3 A0A1B6JNR2 A0A1B6CUY3 A0A2P8YQL9 A0A1B6M5M1 A0A3L8DC02 A0A026WLE0 A0A067RX24 E0VZQ3 A0A2A3EM88 A0A087ZZI7 F4WEW0 A0A151X9X4 E9JCN0 A0A151JNL4 A0A151K1E3 W5JVH8 A0A310SCJ2 A0A154PK42 A0A0J7KYL3 A0A232FL31 A0A0L7QNC7 A0A336MJW4 A0A336L3W0 K7J5G3 A0A195B5U2 A0A158NWX1 A0A0P5DHV1 A0A182FUS3 E9G2Y5 A0A0P4VRU5 A0A0P5HZH7 A0A1Q3F0B8 A0A2J7Q8Y0 A0A0P5C8B0 B0XD44 A0A291S6Y1 A0A0P5B466 A0A087SWY7 A0A1B0CJD7 A0A1J1J5C0 A0A0C9RZJ3 A0JN36 J3S495 A0A2R5LAI8 A0A0P5ZDE8 K7F4Y8 H0VZ66 A0A0P5Z9P4 K7F4Y6 A0A1B0DNX2 Q6AYJ1 G1NLK6 A0A2Y9NEU0 A0A250YKF7 A0A341ADV5 A0A2U4BG13 T1E5X8 G1P5G3 M3XM99 A0A091ELU9 A0A2Y9FPS5 M3WQ95 A0A0B8RW48 A0A287BGJ9 A0A384B6I2 Q8AYS4 F1PNP1 A0A3Q7T7K4 A0A091X1R1 L5KKB3 A0A084VID9 A0A340WP44 A0A3Q7WV68 A0A384C948 A0A2D4PLK1 W5QGJ8 A0A091RGT0 A0A226N6V9 A0A182PC96 A0A1S3FXI9 L8INL4 A0A210PLF8 R0KZK1 A0A2S2QP09 G5BEF7 G1MCS7 F1SR01 S7N182 A0A0A0A487
EC Number
3.6.4.12
2.4.1.11
2.4.1.11
Pubmed
19121390
22118469
26227816
26354079
23674305
29403074
+ More
30249741 24508170 24845553 20566863 21719571 21282665 20920257 23761445 28648823 20075255 21347285 21292972 28992199 19393038 23025625 17381049 21993624 15489334 22673903 20838655 28087693 23758969 17975172 25727380 12724411 16341006 23258410 24438588 24813606 20809919 22751099 28812685 21993625 20010809
30249741 24508170 24845553 20566863 21719571 21282665 20920257 23761445 28648823 20075255 21347285 21292972 28992199 19393038 23025625 17381049 21993624 15489334 22673903 20838655 28087693 23758969 17975172 25727380 12724411 16341006 23258410 24438588 24813606 20809919 22751099 28812685 21993625 20010809
EMBL
BABH01020287
AGBW02008375
OWR53551.1
NWSH01001193
PCG72187.1
JTDY01001185
+ More
KOB74615.1 KQ461155 KPJ08329.1 ODYU01008735 SOQ52592.1 KC469893 AGC92678.1 CU367882 CBH09254.1 KQ459232 KPJ02603.1 HM449906 ADO33041.1 NEVH01016945 PNF25033.1 GECU01006887 JAT00820.1 GEDC01020006 JAS17292.1 PYGN01000430 PSN46523.1 GEBQ01008741 JAT31236.1 QOIP01000010 RLU17995.1 KK107167 EZA56471.1 KK852411 KDR24449.1 DS235854 EEB18859.1 KZ288212 PBC32800.1 GL888111 EGI67277.1 KQ982365 KYQ57144.1 GL771849 EFZ09441.1 KQ978820 KYN28147.1 KQ981189 KYN45238.1 ADMH02000317 ETN67005.1 KQ779356 OAD51993.1 KQ434924 KZC11590.1 LBMM01001888 KMQ95602.1 NNAY01000065 OXU31365.1 KQ414855 KOC60130.1 UFQT01001468 SSX30704.1 UFQS01000967 UFQT01000967 SSX08025.1 SSX28228.1 KQ976595 KYM79640.1 ADTU01028728 GDIP01157272 JAJ66130.1 GL732530 EFX86121.1 GDRN01102836 JAI58169.1 GDIQ01220890 JAK30835.1 GFDL01014045 JAV21000.1 PNF25032.1 GDIP01174467 JAJ48935.1 DS232740 EDS45279.1 MF433010 ATL75361.1 GDIP01189927 JAJ33475.1 KK112335 KFM57376.1 AJWK01014499 CVRI01000070 CRL07154.1 GBYB01002997 GBYB01014855 JAG72764.1 JAG84622.1 BC126495 AAI26496.1 JU174376 AFJ49902.1 GGLE01002387 MBY06513.1 GDIP01046288 JAM57427.1 AGCU01073460 AAKN02031107 GDIP01047888 JAM55827.1 AJVK01007806 AJVK01007807 BC079026 GFFW01000665 JAV44123.1 GAAZ01000710 JAA97233.1 AAPE02021857 AEYP01077487 AEYP01077488 AEYP01077489 KN121399 KFO36571.1 AANG04001751 GBKC01001018 JAG45052.1 AEMK02000034 AB074261 BAC20377.1 AAEX03015200 KK734379 KFR07486.1 KB030666 ELK11985.1 ATLV01013353 KE524854 KFB37733.1 IACN01079963 IACN01079967 LAB58902.1 AMGL01082902 KK817190 KFQ38701.1 MCFN01000170 OXB63336.1 JH880877 ELR58155.1 NEDP02005591 OWF37321.1 KB743435 EOA98763.1 GGMS01010264 MBY79467.1 JH169815 EHB07668.1 ACTA01043018 ACTA01051018 KE162842 EPQ09780.1 KL870627 KGL88871.1
KOB74615.1 KQ461155 KPJ08329.1 ODYU01008735 SOQ52592.1 KC469893 AGC92678.1 CU367882 CBH09254.1 KQ459232 KPJ02603.1 HM449906 ADO33041.1 NEVH01016945 PNF25033.1 GECU01006887 JAT00820.1 GEDC01020006 JAS17292.1 PYGN01000430 PSN46523.1 GEBQ01008741 JAT31236.1 QOIP01000010 RLU17995.1 KK107167 EZA56471.1 KK852411 KDR24449.1 DS235854 EEB18859.1 KZ288212 PBC32800.1 GL888111 EGI67277.1 KQ982365 KYQ57144.1 GL771849 EFZ09441.1 KQ978820 KYN28147.1 KQ981189 KYN45238.1 ADMH02000317 ETN67005.1 KQ779356 OAD51993.1 KQ434924 KZC11590.1 LBMM01001888 KMQ95602.1 NNAY01000065 OXU31365.1 KQ414855 KOC60130.1 UFQT01001468 SSX30704.1 UFQS01000967 UFQT01000967 SSX08025.1 SSX28228.1 KQ976595 KYM79640.1 ADTU01028728 GDIP01157272 JAJ66130.1 GL732530 EFX86121.1 GDRN01102836 JAI58169.1 GDIQ01220890 JAK30835.1 GFDL01014045 JAV21000.1 PNF25032.1 GDIP01174467 JAJ48935.1 DS232740 EDS45279.1 MF433010 ATL75361.1 GDIP01189927 JAJ33475.1 KK112335 KFM57376.1 AJWK01014499 CVRI01000070 CRL07154.1 GBYB01002997 GBYB01014855 JAG72764.1 JAG84622.1 BC126495 AAI26496.1 JU174376 AFJ49902.1 GGLE01002387 MBY06513.1 GDIP01046288 JAM57427.1 AGCU01073460 AAKN02031107 GDIP01047888 JAM55827.1 AJVK01007806 AJVK01007807 BC079026 GFFW01000665 JAV44123.1 GAAZ01000710 JAA97233.1 AAPE02021857 AEYP01077487 AEYP01077488 AEYP01077489 KN121399 KFO36571.1 AANG04001751 GBKC01001018 JAG45052.1 AEMK02000034 AB074261 BAC20377.1 AAEX03015200 KK734379 KFR07486.1 KB030666 ELK11985.1 ATLV01013353 KE524854 KFB37733.1 IACN01079963 IACN01079967 LAB58902.1 AMGL01082902 KK817190 KFQ38701.1 MCFN01000170 OXB63336.1 JH880877 ELR58155.1 NEDP02005591 OWF37321.1 KB743435 EOA98763.1 GGMS01010264 MBY79467.1 JH169815 EHB07668.1 ACTA01043018 ACTA01051018 KE162842 EPQ09780.1 KL870627 KGL88871.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000037510
UP000053240
UP000053268
+ More
UP000235965 UP000245037 UP000279307 UP000053097 UP000027135 UP000009046 UP000242457 UP000005203 UP000007755 UP000075809 UP000078492 UP000078541 UP000000673 UP000076502 UP000036403 UP000215335 UP000053825 UP000002358 UP000078540 UP000005205 UP000069272 UP000000305 UP000002320 UP000054359 UP000092461 UP000183832 UP000009136 UP000007267 UP000005447 UP000092462 UP000002494 UP000001645 UP000248483 UP000252040 UP000245320 UP000001074 UP000000715 UP000028990 UP000248484 UP000011712 UP000008227 UP000261681 UP000002254 UP000286640 UP000053605 UP000010552 UP000030765 UP000265300 UP000286642 UP000261680 UP000291021 UP000002356 UP000198323 UP000075885 UP000081671 UP000242188 UP000006813 UP000008912 UP000053858
UP000235965 UP000245037 UP000279307 UP000053097 UP000027135 UP000009046 UP000242457 UP000005203 UP000007755 UP000075809 UP000078492 UP000078541 UP000000673 UP000076502 UP000036403 UP000215335 UP000053825 UP000002358 UP000078540 UP000005205 UP000069272 UP000000305 UP000002320 UP000054359 UP000092461 UP000183832 UP000009136 UP000007267 UP000005447 UP000092462 UP000002494 UP000001645 UP000248483 UP000252040 UP000245320 UP000001074 UP000000715 UP000028990 UP000248484 UP000011712 UP000008227 UP000261681 UP000002254 UP000286640 UP000053605 UP000010552 UP000030765 UP000265300 UP000286642 UP000261680 UP000291021 UP000002356 UP000198323 UP000075885 UP000081671 UP000242188 UP000006813 UP000008912 UP000053858
Pfam
Interpro
IPR001650
Helicase_C
+ More
IPR036388 WH-like_DNA-bd_sf
IPR014001 Helicase_ATP-bd
IPR011545 DEAD/DEAH_box_helicase_dom
IPR004589 DNA_helicase_ATP-dep_RecQ
IPR032284 RecQ_Zn-bd
IPR027417 P-loop_NTPase
IPR001478 PDZ
IPR036034 PDZ_sf
IPR037213 Run_dom_sf
IPR004012 Run_dom
IPR018982 RQC_domain
IPR008631 Glycogen_synth
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR036388 WH-like_DNA-bd_sf
IPR014001 Helicase_ATP-bd
IPR011545 DEAD/DEAH_box_helicase_dom
IPR004589 DNA_helicase_ATP-dep_RecQ
IPR032284 RecQ_Zn-bd
IPR027417 P-loop_NTPase
IPR001478 PDZ
IPR036034 PDZ_sf
IPR037213 Run_dom_sf
IPR004012 Run_dom
IPR018982 RQC_domain
IPR008631 Glycogen_synth
IPR002464 DNA/RNA_helicase_DEAH_CS
Gene 3D
CDD
ProteinModelPortal
H9J824
A0A212FIK8
A0A2A4JJE6
A0A0L7LGS0
A0A194QSN4
A0A2H1WI67
+ More
L7X1M6 C9S278 A0A194QAP1 E3UKN2 A0A2J7Q8Y3 A0A1B6JNR2 A0A1B6CUY3 A0A2P8YQL9 A0A1B6M5M1 A0A3L8DC02 A0A026WLE0 A0A067RX24 E0VZQ3 A0A2A3EM88 A0A087ZZI7 F4WEW0 A0A151X9X4 E9JCN0 A0A151JNL4 A0A151K1E3 W5JVH8 A0A310SCJ2 A0A154PK42 A0A0J7KYL3 A0A232FL31 A0A0L7QNC7 A0A336MJW4 A0A336L3W0 K7J5G3 A0A195B5U2 A0A158NWX1 A0A0P5DHV1 A0A182FUS3 E9G2Y5 A0A0P4VRU5 A0A0P5HZH7 A0A1Q3F0B8 A0A2J7Q8Y0 A0A0P5C8B0 B0XD44 A0A291S6Y1 A0A0P5B466 A0A087SWY7 A0A1B0CJD7 A0A1J1J5C0 A0A0C9RZJ3 A0JN36 J3S495 A0A2R5LAI8 A0A0P5ZDE8 K7F4Y8 H0VZ66 A0A0P5Z9P4 K7F4Y6 A0A1B0DNX2 Q6AYJ1 G1NLK6 A0A2Y9NEU0 A0A250YKF7 A0A341ADV5 A0A2U4BG13 T1E5X8 G1P5G3 M3XM99 A0A091ELU9 A0A2Y9FPS5 M3WQ95 A0A0B8RW48 A0A287BGJ9 A0A384B6I2 Q8AYS4 F1PNP1 A0A3Q7T7K4 A0A091X1R1 L5KKB3 A0A084VID9 A0A340WP44 A0A3Q7WV68 A0A384C948 A0A2D4PLK1 W5QGJ8 A0A091RGT0 A0A226N6V9 A0A182PC96 A0A1S3FXI9 L8INL4 A0A210PLF8 R0KZK1 A0A2S2QP09 G5BEF7 G1MCS7 F1SR01 S7N182 A0A0A0A487
L7X1M6 C9S278 A0A194QAP1 E3UKN2 A0A2J7Q8Y3 A0A1B6JNR2 A0A1B6CUY3 A0A2P8YQL9 A0A1B6M5M1 A0A3L8DC02 A0A026WLE0 A0A067RX24 E0VZQ3 A0A2A3EM88 A0A087ZZI7 F4WEW0 A0A151X9X4 E9JCN0 A0A151JNL4 A0A151K1E3 W5JVH8 A0A310SCJ2 A0A154PK42 A0A0J7KYL3 A0A232FL31 A0A0L7QNC7 A0A336MJW4 A0A336L3W0 K7J5G3 A0A195B5U2 A0A158NWX1 A0A0P5DHV1 A0A182FUS3 E9G2Y5 A0A0P4VRU5 A0A0P5HZH7 A0A1Q3F0B8 A0A2J7Q8Y0 A0A0P5C8B0 B0XD44 A0A291S6Y1 A0A0P5B466 A0A087SWY7 A0A1B0CJD7 A0A1J1J5C0 A0A0C9RZJ3 A0JN36 J3S495 A0A2R5LAI8 A0A0P5ZDE8 K7F4Y8 H0VZ66 A0A0P5Z9P4 K7F4Y6 A0A1B0DNX2 Q6AYJ1 G1NLK6 A0A2Y9NEU0 A0A250YKF7 A0A341ADV5 A0A2U4BG13 T1E5X8 G1P5G3 M3XM99 A0A091ELU9 A0A2Y9FPS5 M3WQ95 A0A0B8RW48 A0A287BGJ9 A0A384B6I2 Q8AYS4 F1PNP1 A0A3Q7T7K4 A0A091X1R1 L5KKB3 A0A084VID9 A0A340WP44 A0A3Q7WV68 A0A384C948 A0A2D4PLK1 W5QGJ8 A0A091RGT0 A0A226N6V9 A0A182PC96 A0A1S3FXI9 L8INL4 A0A210PLF8 R0KZK1 A0A2S2QP09 G5BEF7 G1MCS7 F1SR01 S7N182 A0A0A0A487
PDB
2WWY
E-value=7.23604e-158,
Score=1431
Ontologies
GO
PANTHER
Topology
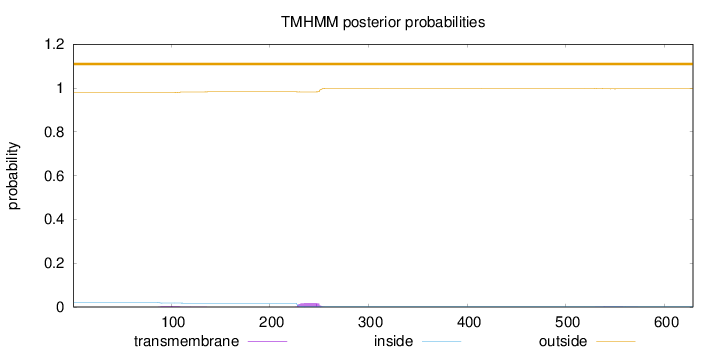
Subcellular location
Nucleus
Length:
629
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.53239
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02099
outside
1 - 629
Population Genetic Test Statistics
Pi
317.447405
Theta
226.777179
Tajima's D
1.119952
CLR
0
CSRT
0.692765361731913
Interpretation
Uncertain
