Gene
KWMTBOMO10090
Pre Gene Modal
BGIBMGA005547
Annotation
PREDICTED:_transcription_factor_Adf-1-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.217
Sequence
CDS
ATGAACGAACTAGACCTGATCAAAGAAGTTGAGAAACGTCCTATACTCTATGATAAATCCGTAAGCGGATTTAACAAAACGAAACTCAGAGATGATGCCTGGAAAGAGGTGCAAGAAGCTCTTAATGTTACAGAATCCGAGTGCAAAAAACGCTGGAGATCTCTGCGTGACTCCTTCATAAAATTACAGCGAACCCATGGAGGTCGTACGAGGTGGCCATATCATCAGGCGATGCGTTTTCTCCTGCCACATATAGAACCTAAGGCGGAAACCGGAATAAAAAAAGAGGAGGATTCCGATCCCGAAGAAGAAGTGGCAAGACAGCGAGCTATACAGAACCTTCCAGACTTCTCATTCAACCATGACAAATACGAAGACGACGAATTTTCAGAGCCATCGCCAAAGAAACAACGTCTCAACTCCACAGATGAAGAATCATGTCAACATTGTACTAGGACGGACCCAGACGAATTATTCTTGCTGAGTTGCGCTCCGACTTTAAAACGGTTGAATTCCAAGAAAAACGCGGTAGCTAGACTCAAGATACAACAAGTGCTTTACGAAGCGGCATTCGGCAGGGAAATGGACGCGTCTTCGGCCCAAGATCACATATATGACGAGCAAGATGATTGA
Protein
MNELDLIKEVEKRPILYDKSVSGFNKTKLRDDAWKEVQEALNVTESECKKRWRSLRDSFIKLQRTHGGRTRWPYHQAMRFLLPHIEPKAETGIKKEEDSDPEEEVARQRAIQNLPDFSFNHDKYEDDEFSEPSPKKQRLNSTDEESCQHCTRTDPDELFLLSCAPTLKRLNSKKNAVARLKIQQVLYEAAFGREMDASSAQDHIYDEQDD
Summary
Uniprot
H9J7Q5
A0A2H1VYR9
A0A212FIC8
A0A2A4JNT6
C9S262
L7X1D7
+ More
A0A194QGL9 A0A194QS45 A0A1W4WQV2 D6WZA0 C4WX15 A0A2S2Q0Z0 A0A1B0G209 A0A1A9UDF4 A0A1B0A611 A0A2H8TP98 A0A1A9WUX6 J9K5V6 A0A1B0BC78 A0A182IZ45 A0A1A9XV79 A0A1I8PUZ1 A0A0L7LE77 A0A1W4X0K5 A0A1B0FHI1 J9KBR0 A0A3B4XFW6 A0A3Q3WAQ6 A0A096M6M6 A0A0P7Y8X8 A0A1A8J5L9 A0A1A8RMH5 A0A1A8BJ41 A0A1A8LKB3 A0A060WUF7 A0A3P9L6E3 A0A1A8UXG3 A0A3B4B9G9 A0A3B4T856 A0A3P9L6D0 A0A3Q2QPZ9 B9ELY0 A0A2A2KXI9 C1BXV3 A0A060WVS3 A0A3P8ZCE2 A0A0P4VMG0 A0A3Q1AN43 A0A2A2L251 A0A3P9DND5 A0A3N0Z5Y5 A0A2I4AP61 A0A3Q1JIN1
A0A194QGL9 A0A194QS45 A0A1W4WQV2 D6WZA0 C4WX15 A0A2S2Q0Z0 A0A1B0G209 A0A1A9UDF4 A0A1B0A611 A0A2H8TP98 A0A1A9WUX6 J9K5V6 A0A1B0BC78 A0A182IZ45 A0A1A9XV79 A0A1I8PUZ1 A0A0L7LE77 A0A1W4X0K5 A0A1B0FHI1 J9KBR0 A0A3B4XFW6 A0A3Q3WAQ6 A0A096M6M6 A0A0P7Y8X8 A0A1A8J5L9 A0A1A8RMH5 A0A1A8BJ41 A0A1A8LKB3 A0A060WUF7 A0A3P9L6E3 A0A1A8UXG3 A0A3B4B9G9 A0A3B4T856 A0A3P9L6D0 A0A3Q2QPZ9 B9ELY0 A0A2A2KXI9 C1BXV3 A0A060WVS3 A0A3P8ZCE2 A0A0P4VMG0 A0A3Q1AN43 A0A2A2L251 A0A3P9DND5 A0A3N0Z5Y5 A0A2I4AP61 A0A3Q1JIN1
Pubmed
EMBL
BABH01020287
BABH01020288
BABH01020289
BABH01020290
BABH01020291
ODYU01005286
+ More
SOQ45989.1 AGBW02008409 OWR53470.1 NWSH01000985 PCG73244.1 CT955980 CBH09242.1 KC469893 AGC92666.1 KQ459232 KPJ02591.1 KQ461155 KPJ08343.1 KQ971372 EFA09743.1 AK342392 BAH72435.1 GGMS01002232 MBY71435.1 CCAG010006187 GFXV01003263 MBW15068.1 ABLF02029435 JXJN01011881 JTDY01001509 KOB73695.1 CCAG010019481 ABLF02017748 ABLF02030228 ABLF02033162 ABLF02055684 ABLF02061509 AYCK01001094 JARO02008890 KPP62202.1 HAED01018472 SBR04917.1 HAEH01018199 SBS07320.1 HADZ01003278 SBP67219.1 HAEF01006957 SBR44339.1 FR904638 CDQ68699.1 HADY01009641 HAEJ01011814 SBS52271.1 BT056655 ACM08527.1 LIAE01007525 PAV78716.1 BT079432 ACO13856.1 FR904771 CDQ71488.1 GDKW01003728 JAI52867.1 LIAE01007297 PAV80248.1 RJVU01007700 ROL53880.1
SOQ45989.1 AGBW02008409 OWR53470.1 NWSH01000985 PCG73244.1 CT955980 CBH09242.1 KC469893 AGC92666.1 KQ459232 KPJ02591.1 KQ461155 KPJ08343.1 KQ971372 EFA09743.1 AK342392 BAH72435.1 GGMS01002232 MBY71435.1 CCAG010006187 GFXV01003263 MBW15068.1 ABLF02029435 JXJN01011881 JTDY01001509 KOB73695.1 CCAG010019481 ABLF02017748 ABLF02030228 ABLF02033162 ABLF02055684 ABLF02061509 AYCK01001094 JARO02008890 KPP62202.1 HAED01018472 SBR04917.1 HAEH01018199 SBS07320.1 HADZ01003278 SBP67219.1 HAEF01006957 SBR44339.1 FR904638 CDQ68699.1 HADY01009641 HAEJ01011814 SBS52271.1 BT056655 ACM08527.1 LIAE01007525 PAV78716.1 BT079432 ACO13856.1 FR904771 CDQ71488.1 GDKW01003728 JAI52867.1 LIAE01007297 PAV80248.1 RJVU01007700 ROL53880.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000192223
+ More
UP000007266 UP000092444 UP000078200 UP000092445 UP000091820 UP000007819 UP000092460 UP000075880 UP000092443 UP000095300 UP000037510 UP000261360 UP000261620 UP000028760 UP000034805 UP000193380 UP000265180 UP000261520 UP000261420 UP000265000 UP000087266 UP000218231 UP000265140 UP000257160 UP000265160 UP000192220 UP000265040
UP000007266 UP000092444 UP000078200 UP000092445 UP000091820 UP000007819 UP000092460 UP000075880 UP000092443 UP000095300 UP000037510 UP000261360 UP000261620 UP000028760 UP000034805 UP000193380 UP000265180 UP000261520 UP000261420 UP000265000 UP000087266 UP000218231 UP000265140 UP000257160 UP000265160 UP000192220 UP000265040
PRIDE
Interpro
SUPFAM
SSF53098
SSF53098
Gene 3D
CDD
ProteinModelPortal
H9J7Q5
A0A2H1VYR9
A0A212FIC8
A0A2A4JNT6
C9S262
L7X1D7
+ More
A0A194QGL9 A0A194QS45 A0A1W4WQV2 D6WZA0 C4WX15 A0A2S2Q0Z0 A0A1B0G209 A0A1A9UDF4 A0A1B0A611 A0A2H8TP98 A0A1A9WUX6 J9K5V6 A0A1B0BC78 A0A182IZ45 A0A1A9XV79 A0A1I8PUZ1 A0A0L7LE77 A0A1W4X0K5 A0A1B0FHI1 J9KBR0 A0A3B4XFW6 A0A3Q3WAQ6 A0A096M6M6 A0A0P7Y8X8 A0A1A8J5L9 A0A1A8RMH5 A0A1A8BJ41 A0A1A8LKB3 A0A060WUF7 A0A3P9L6E3 A0A1A8UXG3 A0A3B4B9G9 A0A3B4T856 A0A3P9L6D0 A0A3Q2QPZ9 B9ELY0 A0A2A2KXI9 C1BXV3 A0A060WVS3 A0A3P8ZCE2 A0A0P4VMG0 A0A3Q1AN43 A0A2A2L251 A0A3P9DND5 A0A3N0Z5Y5 A0A2I4AP61 A0A3Q1JIN1
A0A194QGL9 A0A194QS45 A0A1W4WQV2 D6WZA0 C4WX15 A0A2S2Q0Z0 A0A1B0G209 A0A1A9UDF4 A0A1B0A611 A0A2H8TP98 A0A1A9WUX6 J9K5V6 A0A1B0BC78 A0A182IZ45 A0A1A9XV79 A0A1I8PUZ1 A0A0L7LE77 A0A1W4X0K5 A0A1B0FHI1 J9KBR0 A0A3B4XFW6 A0A3Q3WAQ6 A0A096M6M6 A0A0P7Y8X8 A0A1A8J5L9 A0A1A8RMH5 A0A1A8BJ41 A0A1A8LKB3 A0A060WUF7 A0A3P9L6E3 A0A1A8UXG3 A0A3B4B9G9 A0A3B4T856 A0A3P9L6D0 A0A3Q2QPZ9 B9ELY0 A0A2A2KXI9 C1BXV3 A0A060WVS3 A0A3P8ZCE2 A0A0P4VMG0 A0A3Q1AN43 A0A2A2L251 A0A3P9DND5 A0A3N0Z5Y5 A0A2I4AP61 A0A3Q1JIN1
Ontologies
GO
PANTHER
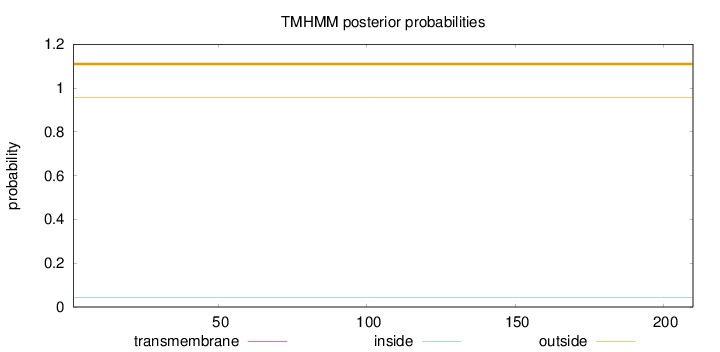
Topology
Subcellular location
Nucleus
Length:
210
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04206
outside
1 - 210
Population Genetic Test Statistics
Pi
167.379534
Theta
150.958315
Tajima's D
0.112052
CLR
1.46736
CSRT
0.407129643517824
Interpretation
Uncertain
