Gene
KWMTBOMO10082
Annotation
PREDICTED:_galactokinase-like_isoform_X1_[Bombyx_mori]
Full name
Galactokinase
Alternative Name
Galactose kinase
Location in the cell
Cytoplasmic Reliability : 2.053
Sequence
CDS
ATGAGGCTGAGAGATCTCGGTTGTAACGAGTTAGTTATCAGAAGAGCTCGGCATGTCATAGGAGAAATTCAGAGAACGGTGGAAGCTGCCATCAAATTGACCGAGAAGAATTACAAAAGGGTGGGTGAACTTTTTTACCAATCGCATGACTCGCTAAGCAAATTGATGGAGGTATCCTGCGTGGAGCTCGATCAGATAGTGGACATCCTCCGGGATGCTCCAGGGGTGTATGGTGCGAGAATGACGGGAGGTGGCTTCGGTGGATGTGCGGTGGCTTTGGTTCAAAAGAGCGAATTGGATGCTTTGAAACAGCGGATTCTGTCGCAATATAAAGGAAAGCCAGTATTTTTCGTGTGCGTACCGAGCGAAGGCGCGAGATTTTTGAAACTTAACCACTGA
Protein
MRLRDLGCNELVIRRARHVIGEIQRTVEAAIKLTEKNYKRVGELFYQSHDSLSKLMEVSCVELDQIVDILRDAPGVYGARMTGGGFGGCAVALVQKSELDALKQRILSQYKGKPVFFVCVPSEGARFLKLNH
Summary
Catalytic Activity
alpha-D-galactose + ATP = ADP + alpha-D-galactose 1-phosphate + H(+)
Subunit
Homodimer.
Similarity
Belongs to the GHMP kinase family. GalK subfamily.
Keywords
ATP-binding
Carbohydrate metabolism
Complete proteome
Galactose metabolism
Kinase
Nucleotide-binding
Phosphoprotein
Reference proteome
Transferase
Feature
chain Galactokinase
Uniprot
A0A194QS49
A0A2A4JN41
A0A2H1WK86
A0A194QC70
A0A212F634
L7X3T9
+ More
A0A3S2NQJ0 A0A1B6FCF7 A0A1W4XGC4 A0A0T6AWY9 A0A1B6IUY6 A0A0S7LS86 U4US63 J3JTL8 A0A3B5LZ96 A0A1A8KMR8 A0A1B6EPN4 A0A3B3YP76 A0A087YH87 J9K0D5 V4A9N7 A0A1A8R1L2 A0A3Q3GZ61 A0A0S7LSX8 A0A210QGH3 A0A1A8I8N9 A0A1V6GMS4 M3ZZJ4 A0A3D3DGP7 A0A3B4ZJN8 A0A1A8CXS4 A0A2H8TKY3 A0A1B0G514 A0A1A8GGH3 A0A3B3VNT8 A0A1A8UAG4 A0A1A8SC52 A0A1A8M8L9 A0A1B6DN88 A0A3Q1JHI9 W5N0D3 G3PRC4 F6X9G1 A0A3P9MWP7 A0A3M6UW61 A0A1A8AXC1 A0A1A8FSR3 T0MDL9 A0A1A9VBU5 A0A224XQA0 H3CAN5 A0A3Q3STN9 A0A3Q2XT66 W5PIM8 D6WVS7 A0A1S3NEQ2 W4XRV9 A0A1W5AWA4 U5EZ50 A0A0L8HES9 F1R3T4 A0A0P7ULY4 A0A3Q1BRE1 A0A3P8TNM3 Q0VFQ1 F7EJ79 A0A2U3X9E9 K7JAZ0 F1RVY7 A0A384AEI6 H0VLY7 A0A2Y9PEF1 V5GG13 G1K1R6 A0A232F4K3 A0A341CPI5 A6H768 A0A3P8WF92 A0A3P4RY69 A0A3Q1G1Q5 L9KZB3 A0A091DYS7 D3TL00 A0A340X518 A0A336LE10 A0A3Q0DKB3 A0A2B4SUG3 H2T403 A0A026WMY5 A0A2Y9ECV8 F7DHW2 L5KLD5 A0A3Q7PIP6 H2YGD8 A0A2U3VI60 A0A3Q2GA27 A0A1J1ICY3 A0A2I0TRT1 A0A0P4VXJ8 R4G3I8
A0A3S2NQJ0 A0A1B6FCF7 A0A1W4XGC4 A0A0T6AWY9 A0A1B6IUY6 A0A0S7LS86 U4US63 J3JTL8 A0A3B5LZ96 A0A1A8KMR8 A0A1B6EPN4 A0A3B3YP76 A0A087YH87 J9K0D5 V4A9N7 A0A1A8R1L2 A0A3Q3GZ61 A0A0S7LSX8 A0A210QGH3 A0A1A8I8N9 A0A1V6GMS4 M3ZZJ4 A0A3D3DGP7 A0A3B4ZJN8 A0A1A8CXS4 A0A2H8TKY3 A0A1B0G514 A0A1A8GGH3 A0A3B3VNT8 A0A1A8UAG4 A0A1A8SC52 A0A1A8M8L9 A0A1B6DN88 A0A3Q1JHI9 W5N0D3 G3PRC4 F6X9G1 A0A3P9MWP7 A0A3M6UW61 A0A1A8AXC1 A0A1A8FSR3 T0MDL9 A0A1A9VBU5 A0A224XQA0 H3CAN5 A0A3Q3STN9 A0A3Q2XT66 W5PIM8 D6WVS7 A0A1S3NEQ2 W4XRV9 A0A1W5AWA4 U5EZ50 A0A0L8HES9 F1R3T4 A0A0P7ULY4 A0A3Q1BRE1 A0A3P8TNM3 Q0VFQ1 F7EJ79 A0A2U3X9E9 K7JAZ0 F1RVY7 A0A384AEI6 H0VLY7 A0A2Y9PEF1 V5GG13 G1K1R6 A0A232F4K3 A0A341CPI5 A6H768 A0A3P8WF92 A0A3P4RY69 A0A3Q1G1Q5 L9KZB3 A0A091DYS7 D3TL00 A0A340X518 A0A336LE10 A0A3Q0DKB3 A0A2B4SUG3 H2T403 A0A026WMY5 A0A2Y9ECV8 F7DHW2 L5KLD5 A0A3Q7PIP6 H2YGD8 A0A2U3VI60 A0A3Q2GA27 A0A1J1ICY3 A0A2I0TRT1 A0A0P4VXJ8 R4G3I8
EC Number
2.7.1.6
Pubmed
26354079
22118469
23674305
23537049
22516182
23254933
+ More
28812685 23542700 30148503 12481130 15114417 30382153 23149746 15496914 20809919 18362917 19820115 23594743 20431018 20075255 30723633 21993624 25765539 19393038 28648823 16305752 24487278 23385571 20353571 21551351 24508170 30249741 19892987 23258410 27129103
28812685 23542700 30148503 12481130 15114417 30382153 23149746 15496914 20809919 18362917 19820115 23594743 20431018 20075255 30723633 21993624 25765539 19393038 28648823 16305752 24487278 23385571 20353571 21551351 24508170 30249741 19892987 23258410 27129103
EMBL
KQ461155
KPJ08348.1
NWSH01000985
PCG73239.1
ODYU01009225
SOQ53493.1
+ More
KQ459232 KPJ02585.1 AGBW02010097 OWR49169.1 KC469893 AGC92660.1 RSAL01000145 RVE45937.1 GECZ01022136 JAS47633.1 LJIG01022605 KRT79673.1 GECU01016968 JAS90738.1 GBYX01152283 JAO92269.1 KB632424 ERL95343.1 APGK01040016 APGK01040017 BT126575 KB740975 AEE61539.1 ENN76513.1 HAEE01013670 SBR33720.1 GECZ01029881 JAS39888.1 AYCK01026357 AYCK01026358 ABLF02008254 ABLF02008255 KB202518 ESO90001.1 HAEH01014182 SBR99338.1 GBYX01152285 JAO92267.1 NEDP02003775 OWF47799.1 HAED01007649 SBQ93861.1 MWEL01000003 OQC27210.1 DPXU01000145 HCL91961.1 HADZ01019694 SBP83635.1 GFXV01002143 MBW13948.1 CCAG010014030 HAEC01002054 SBQ70131.1 HAEJ01004648 SBS45105.1 HAEI01013437 SBS15906.1 HAEF01011752 HAEG01010736 SBR52911.1 GEDC01029996 GEDC01020334 GEDC01014251 GEDC01014058 GEDC01010160 GEDC01002359 JAS07302.1 JAS16964.1 JAS23047.1 JAS23240.1 JAS27138.1 JAS34939.1 AHAT01000285 EAAA01001823 RCHS01000645 RMX57578.1 HADY01020390 SBP58875.1 HAEB01015279 SBQ61806.1 KB016558 EQB77251.1 GFTR01005796 JAW10630.1 AMGL01019310 KQ971358 EFA08251.1 AAGJ04033376 GANO01001429 JAB58442.1 KQ418322 KOF87771.1 FQ312018 JARO02008297 KPP62899.1 BC118744 AAI18745.1 AAMC01098331 AAMC01098332 AAMC01098333 AAMC01098334 AAMC01098335 AAZX01004191 AAZX01015652 AEMK02000080 DQIR01091093 DQIR01097081 DQIR01223483 DQIR01228524 DQIR01245916 DQIR01245917 DQIR01291019 DQIR01292230 DQIR01312380 HDA46569.1 AAKN02056017 GANP01015323 JAB69145.1 DAAA02049512 DAAA02049513 DAAA02049514 NNAY01001063 OXU25277.1 BC146132 BT025341 BT025347 BT025488 BT026290 CYRY02044295 VCX39175.1 KB320577 ELW68310.1 KN121670 KFO35618.1 EZ422090 ADD18295.1 UFQS01002974 UFQT01002974 SSX14997.1 SSX34383.1 LSMT01000028 PFX32035.1 KK107167 QOIP01000010 EZA56469.1 RLU18001.1 KB030661 ELK12360.1 CVRI01000047 CRK97610.1 KZ507598 PKU36499.1 GDKW01002282 JAI54313.1 ACPB03022326 GAHY01001641 JAA75869.1
KQ459232 KPJ02585.1 AGBW02010097 OWR49169.1 KC469893 AGC92660.1 RSAL01000145 RVE45937.1 GECZ01022136 JAS47633.1 LJIG01022605 KRT79673.1 GECU01016968 JAS90738.1 GBYX01152283 JAO92269.1 KB632424 ERL95343.1 APGK01040016 APGK01040017 BT126575 KB740975 AEE61539.1 ENN76513.1 HAEE01013670 SBR33720.1 GECZ01029881 JAS39888.1 AYCK01026357 AYCK01026358 ABLF02008254 ABLF02008255 KB202518 ESO90001.1 HAEH01014182 SBR99338.1 GBYX01152285 JAO92267.1 NEDP02003775 OWF47799.1 HAED01007649 SBQ93861.1 MWEL01000003 OQC27210.1 DPXU01000145 HCL91961.1 HADZ01019694 SBP83635.1 GFXV01002143 MBW13948.1 CCAG010014030 HAEC01002054 SBQ70131.1 HAEJ01004648 SBS45105.1 HAEI01013437 SBS15906.1 HAEF01011752 HAEG01010736 SBR52911.1 GEDC01029996 GEDC01020334 GEDC01014251 GEDC01014058 GEDC01010160 GEDC01002359 JAS07302.1 JAS16964.1 JAS23047.1 JAS23240.1 JAS27138.1 JAS34939.1 AHAT01000285 EAAA01001823 RCHS01000645 RMX57578.1 HADY01020390 SBP58875.1 HAEB01015279 SBQ61806.1 KB016558 EQB77251.1 GFTR01005796 JAW10630.1 AMGL01019310 KQ971358 EFA08251.1 AAGJ04033376 GANO01001429 JAB58442.1 KQ418322 KOF87771.1 FQ312018 JARO02008297 KPP62899.1 BC118744 AAI18745.1 AAMC01098331 AAMC01098332 AAMC01098333 AAMC01098334 AAMC01098335 AAZX01004191 AAZX01015652 AEMK02000080 DQIR01091093 DQIR01097081 DQIR01223483 DQIR01228524 DQIR01245916 DQIR01245917 DQIR01291019 DQIR01292230 DQIR01312380 HDA46569.1 AAKN02056017 GANP01015323 JAB69145.1 DAAA02049512 DAAA02049513 DAAA02049514 NNAY01001063 OXU25277.1 BC146132 BT025341 BT025347 BT025488 BT026290 CYRY02044295 VCX39175.1 KB320577 ELW68310.1 KN121670 KFO35618.1 EZ422090 ADD18295.1 UFQS01002974 UFQT01002974 SSX14997.1 SSX34383.1 LSMT01000028 PFX32035.1 KK107167 QOIP01000010 EZA56469.1 RLU18001.1 KB030661 ELK12360.1 CVRI01000047 CRK97610.1 KZ507598 PKU36499.1 GDKW01002282 JAI54313.1 ACPB03022326 GAHY01001641 JAA75869.1
Proteomes
UP000053240
UP000218220
UP000053268
UP000007151
UP000283053
UP000192223
+ More
UP000030742 UP000019118 UP000261380 UP000261480 UP000028760 UP000007819 UP000030746 UP000261660 UP000242188 UP000002852 UP000261400 UP000092444 UP000261500 UP000265040 UP000018468 UP000007635 UP000008144 UP000242638 UP000275408 UP000078200 UP000007303 UP000261640 UP000264820 UP000002356 UP000007266 UP000087266 UP000007110 UP000192224 UP000053454 UP000000437 UP000034805 UP000257160 UP000265080 UP000008143 UP000245341 UP000002358 UP000008227 UP000261681 UP000005447 UP000248483 UP000009136 UP000215335 UP000252040 UP000265120 UP000257200 UP000011518 UP000028990 UP000265300 UP000189704 UP000225706 UP000005226 UP000053097 UP000279307 UP000002281 UP000010552 UP000286641 UP000007875 UP000245340 UP000265020 UP000183832 UP000015103
UP000030742 UP000019118 UP000261380 UP000261480 UP000028760 UP000007819 UP000030746 UP000261660 UP000242188 UP000002852 UP000261400 UP000092444 UP000261500 UP000265040 UP000018468 UP000007635 UP000008144 UP000242638 UP000275408 UP000078200 UP000007303 UP000261640 UP000264820 UP000002356 UP000007266 UP000087266 UP000007110 UP000192224 UP000053454 UP000000437 UP000034805 UP000257160 UP000265080 UP000008143 UP000245341 UP000002358 UP000008227 UP000261681 UP000005447 UP000248483 UP000009136 UP000215335 UP000252040 UP000265120 UP000257200 UP000011518 UP000028990 UP000265300 UP000189704 UP000225706 UP000005226 UP000053097 UP000279307 UP000002281 UP000010552 UP000286641 UP000007875 UP000245340 UP000265020 UP000183832 UP000015103
Interpro
IPR006206
Mevalonate/galactokinase
+ More
IPR013750 GHMP_kinase_C_dom
IPR019741 Galactokinase_CS
IPR036554 GHMP_kinase_C_sf
IPR006204 GHMP_kinase_N_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR019539 GalKase_gal-bd
IPR020568 Ribosomal_S5_D2-typ_fold
IPR000705 Galactokinase
IPR006203 GHMP_knse_ATP-bd_CS
IPR013750 GHMP_kinase_C_dom
IPR019741 Galactokinase_CS
IPR036554 GHMP_kinase_C_sf
IPR006204 GHMP_kinase_N_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR019539 GalKase_gal-bd
IPR020568 Ribosomal_S5_D2-typ_fold
IPR000705 Galactokinase
IPR006203 GHMP_knse_ATP-bd_CS
Gene 3D
ProteinModelPortal
A0A194QS49
A0A2A4JN41
A0A2H1WK86
A0A194QC70
A0A212F634
L7X3T9
+ More
A0A3S2NQJ0 A0A1B6FCF7 A0A1W4XGC4 A0A0T6AWY9 A0A1B6IUY6 A0A0S7LS86 U4US63 J3JTL8 A0A3B5LZ96 A0A1A8KMR8 A0A1B6EPN4 A0A3B3YP76 A0A087YH87 J9K0D5 V4A9N7 A0A1A8R1L2 A0A3Q3GZ61 A0A0S7LSX8 A0A210QGH3 A0A1A8I8N9 A0A1V6GMS4 M3ZZJ4 A0A3D3DGP7 A0A3B4ZJN8 A0A1A8CXS4 A0A2H8TKY3 A0A1B0G514 A0A1A8GGH3 A0A3B3VNT8 A0A1A8UAG4 A0A1A8SC52 A0A1A8M8L9 A0A1B6DN88 A0A3Q1JHI9 W5N0D3 G3PRC4 F6X9G1 A0A3P9MWP7 A0A3M6UW61 A0A1A8AXC1 A0A1A8FSR3 T0MDL9 A0A1A9VBU5 A0A224XQA0 H3CAN5 A0A3Q3STN9 A0A3Q2XT66 W5PIM8 D6WVS7 A0A1S3NEQ2 W4XRV9 A0A1W5AWA4 U5EZ50 A0A0L8HES9 F1R3T4 A0A0P7ULY4 A0A3Q1BRE1 A0A3P8TNM3 Q0VFQ1 F7EJ79 A0A2U3X9E9 K7JAZ0 F1RVY7 A0A384AEI6 H0VLY7 A0A2Y9PEF1 V5GG13 G1K1R6 A0A232F4K3 A0A341CPI5 A6H768 A0A3P8WF92 A0A3P4RY69 A0A3Q1G1Q5 L9KZB3 A0A091DYS7 D3TL00 A0A340X518 A0A336LE10 A0A3Q0DKB3 A0A2B4SUG3 H2T403 A0A026WMY5 A0A2Y9ECV8 F7DHW2 L5KLD5 A0A3Q7PIP6 H2YGD8 A0A2U3VI60 A0A3Q2GA27 A0A1J1ICY3 A0A2I0TRT1 A0A0P4VXJ8 R4G3I8
A0A3S2NQJ0 A0A1B6FCF7 A0A1W4XGC4 A0A0T6AWY9 A0A1B6IUY6 A0A0S7LS86 U4US63 J3JTL8 A0A3B5LZ96 A0A1A8KMR8 A0A1B6EPN4 A0A3B3YP76 A0A087YH87 J9K0D5 V4A9N7 A0A1A8R1L2 A0A3Q3GZ61 A0A0S7LSX8 A0A210QGH3 A0A1A8I8N9 A0A1V6GMS4 M3ZZJ4 A0A3D3DGP7 A0A3B4ZJN8 A0A1A8CXS4 A0A2H8TKY3 A0A1B0G514 A0A1A8GGH3 A0A3B3VNT8 A0A1A8UAG4 A0A1A8SC52 A0A1A8M8L9 A0A1B6DN88 A0A3Q1JHI9 W5N0D3 G3PRC4 F6X9G1 A0A3P9MWP7 A0A3M6UW61 A0A1A8AXC1 A0A1A8FSR3 T0MDL9 A0A1A9VBU5 A0A224XQA0 H3CAN5 A0A3Q3STN9 A0A3Q2XT66 W5PIM8 D6WVS7 A0A1S3NEQ2 W4XRV9 A0A1W5AWA4 U5EZ50 A0A0L8HES9 F1R3T4 A0A0P7ULY4 A0A3Q1BRE1 A0A3P8TNM3 Q0VFQ1 F7EJ79 A0A2U3X9E9 K7JAZ0 F1RVY7 A0A384AEI6 H0VLY7 A0A2Y9PEF1 V5GG13 G1K1R6 A0A232F4K3 A0A341CPI5 A6H768 A0A3P8WF92 A0A3P4RY69 A0A3Q1G1Q5 L9KZB3 A0A091DYS7 D3TL00 A0A340X518 A0A336LE10 A0A3Q0DKB3 A0A2B4SUG3 H2T403 A0A026WMY5 A0A2Y9ECV8 F7DHW2 L5KLD5 A0A3Q7PIP6 H2YGD8 A0A2U3VI60 A0A3Q2GA27 A0A1J1ICY3 A0A2I0TRT1 A0A0P4VXJ8 R4G3I8
PDB
6QJE
E-value=1.87903e-25,
Score=280
Ontologies
GO
PANTHER
Topology
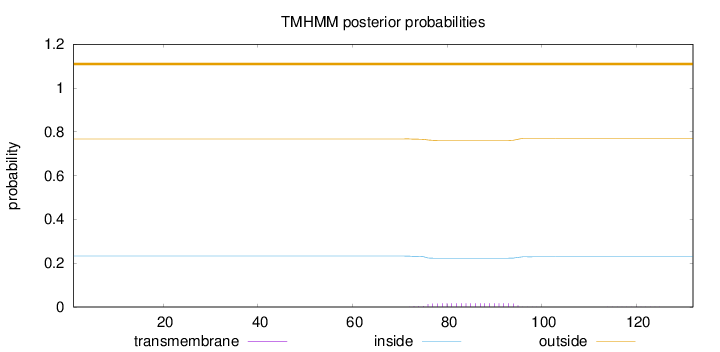
Length:
132
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.32438
Exp number, first 60 AAs:
0
Total prob of N-in:
0.23237
outside
1 - 132
Population Genetic Test Statistics
Pi
197.989284
Theta
146.70236
Tajima's D
0.551808
CLR
0.344763
CSRT
0.527573621318934
Interpretation
Uncertain
