Gene
KWMTBOMO10078
Annotation
PREDICTED:_guanylate_cyclase_32E_[Amyelois_transitella]
Full name
Guanylate cyclase
+ More
Guanylate cyclase 32E
Guanylate cyclase 32E
Location in the cell
Extracellular Reliability : 1.623 PlasmaMembrane Reliability : 1.112
Sequence
CDS
ATGTTTTTTGTTACAGTATACGTGTTCCTGGGCGAACACATAGCGCTTGTCGACTTCGTAAACGTGCTGGAGAAACGGGGATTGCTCGCTACAGGAAACTATGCAGTTGTCGCTGTCGACGATGAGATCTATGACCCTAACACCACAGCCGTGACTTATGCTGATCATCTCGGAAAAAACCGCACCAACGACATAACTGCGTTCAGGTCTGTCCTGAGACTCACGCCTTCGTTCCCAACGAATCCTCATTACGAGATCCTTTGTCAAATGATCAGGGAGCTCTCGGCCGTGCCACCATTCTGCGTTCCGAACTATCACAAGATCTTCGAAGATGCCTCGGGATAA
Protein
MFFVTVYVFLGEHIALVDFVNVLEKRGLLATGNYAVVAVDDEIYDPNTTAVTYADHLGKNRTNDITAFRSVLRLTPSFPTNPHYEILCQMIRELSAVPPFCVPNYHKIFEDASG
Summary
Catalytic Activity
GTP = 3',5'-cyclic GMP + diphosphate
Similarity
Belongs to the adenylyl cyclase class-4/guanylyl cyclase family.
Keywords
cGMP biosynthesis
Complete proteome
Disulfide bond
Glycoprotein
GTP-binding
Lyase
Membrane
Nucleotide-binding
Receptor
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Guanylate cyclase
Uniprot
A0A194QSQ1
A0A194QAM5
A0A2A4JP88
A0A139WDI2
A0A139WDK0
T1HNK9
+ More
A0A146KWY4 A0A067RMP4 A0A2P8XXW2 E0W2G4 B4LQH0 B4KHA8 N6TFV7 A0A1B0D0D2 A0A0N8E865 A0A0P5CQL6 A0A0P5CN85 A0A0N8CLV3 A0A0P5WH20 A0A0P6BHS8 A0A0P5EGZ5 A0A0P4YXA1 A0A0P6G379 A0A0N8A6H0 A0A0P4ZRW1 A0A0P5WUA0 A0A0P5B5W5 A0A0P5UHG3 A0A0P6HWV4 A0A0P6G382 A0A0P5DR45 A0A164Q4I2 A0A0P5DGF2 A0A0P5D5R6 A0A0P5VEM9 A0A2R7WWS8 A0A3B0JFK9 B5DIL1 A0A0R3NYC5 A0A0R1DJU5 A0A0R3NTK6 A0A1A9UXC2 A0A1B0BWX4 A0A1B0A0T9 A0A0J9R1J4 A0A1B0FQA5 A0A1W4W5P4 A0A0Q5WB16 A8DYZ3 B3MMW4 B4MWE1 A0A1I8PD95 A0A084VYH2 A0A182F1Z3 W5JRT6 A0A182ILP4 A0A0L0BZE8 A0A182Y970 A0A182Q3F5 A0A182NP43 A0A182R498 A0A0P5CS31 A0A1I8NG04 A0A182VBW3 A0A182VU62 A0A1S4H063 A0A182XDI7 A0A2C9GRZ6 A0A182U4U3 A0A182KNH9 A0A0M4E8L4 Q7PWU5 B0WPU2 A0A1S4FAA0 A0A182H963 A0A0P5LTI7 A0A0P5QU44 A0A0P6FWN6 A0A0P5PAZ8 A0A0P5ZAY1 A0A0P5DI19 A0A0P5RTM9 A0A0P6DLI3 A0A0P6B8G8 A0A0N8BNB3 A0A0N8EM03 A0A0P5NXS8 A0A182P9Q0 A0A182SNN7 A0A1A9Y362 B4P180 B4HX26 B4QAC0 B3N4K3 Q07553 Q17AF9 B4G9U6 B4JBD1 A0A162R3A0
A0A146KWY4 A0A067RMP4 A0A2P8XXW2 E0W2G4 B4LQH0 B4KHA8 N6TFV7 A0A1B0D0D2 A0A0N8E865 A0A0P5CQL6 A0A0P5CN85 A0A0N8CLV3 A0A0P5WH20 A0A0P6BHS8 A0A0P5EGZ5 A0A0P4YXA1 A0A0P6G379 A0A0N8A6H0 A0A0P4ZRW1 A0A0P5WUA0 A0A0P5B5W5 A0A0P5UHG3 A0A0P6HWV4 A0A0P6G382 A0A0P5DR45 A0A164Q4I2 A0A0P5DGF2 A0A0P5D5R6 A0A0P5VEM9 A0A2R7WWS8 A0A3B0JFK9 B5DIL1 A0A0R3NYC5 A0A0R1DJU5 A0A0R3NTK6 A0A1A9UXC2 A0A1B0BWX4 A0A1B0A0T9 A0A0J9R1J4 A0A1B0FQA5 A0A1W4W5P4 A0A0Q5WB16 A8DYZ3 B3MMW4 B4MWE1 A0A1I8PD95 A0A084VYH2 A0A182F1Z3 W5JRT6 A0A182ILP4 A0A0L0BZE8 A0A182Y970 A0A182Q3F5 A0A182NP43 A0A182R498 A0A0P5CS31 A0A1I8NG04 A0A182VBW3 A0A182VU62 A0A1S4H063 A0A182XDI7 A0A2C9GRZ6 A0A182U4U3 A0A182KNH9 A0A0M4E8L4 Q7PWU5 B0WPU2 A0A1S4FAA0 A0A182H963 A0A0P5LTI7 A0A0P5QU44 A0A0P6FWN6 A0A0P5PAZ8 A0A0P5ZAY1 A0A0P5DI19 A0A0P5RTM9 A0A0P6DLI3 A0A0P6B8G8 A0A0N8BNB3 A0A0N8EM03 A0A0P5NXS8 A0A182P9Q0 A0A182SNN7 A0A1A9Y362 B4P180 B4HX26 B4QAC0 B3N4K3 Q07553 Q17AF9 B4G9U6 B4JBD1 A0A162R3A0
EC Number
4.6.1.2
Pubmed
EMBL
KQ461155
KPJ08349.1
KQ459232
KPJ02583.1
NWSH01000900
PCG73629.1
+ More
KQ971357 KYB26028.1 KYB26029.1 ACPB03001838 GDHC01017635 JAQ00994.1 KK852421 KDR24308.1 PYGN01001181 PSN36852.1 DS235878 EEB19820.1 CH940649 EDW63420.2 KRF81107.1 CH933807 EDW13325.2 APGK01039614 APGK01039615 APGK01039616 KB740972 ENN76628.1 AJVK01002401 AJVK01002402 AJVK01002403 AJVK01002404 GDIQ01052348 JAN42389.1 GDIP01166914 JAJ56488.1 GDIP01167810 JAJ55592.1 GDIP01119023 JAL84691.1 GDIP01086478 JAM17237.1 GDIP01014364 JAM89351.1 GDIP01160124 JAJ63278.1 GDIP01222214 JAJ01188.1 GDIQ01039146 JAN55591.1 GDIP01177777 JAJ45625.1 GDIP01222213 GDIP01222212 JAJ01189.1 GDIP01081672 JAM22043.1 GDIP01189168 JAJ34234.1 GDIP01112873 JAL90841.1 GDIQ01014350 JAN80387.1 GDIQ01039147 JAN55590.1 GDIP01158720 JAJ64682.1 LRGB01002475 KZS07425.1 GDIP01160123 JAJ63279.1 GDIP01160822 JAJ62580.1 GDIP01102876 JAM00839.1 KK855823 PTY23999.1 OUUW01000004 SPP79483.1 CH379060 EDY70153.2 KRT04269.1 CM000157 KRJ97540.1 KRT04268.1 JXJN01021989 JXJN01021990 CM002910 KMY89709.1 CCAG010004953 CH954177 KQS70572.1 AE014134 ABV53670.1 CH902620 EDV30989.2 CH963857 EDW76011.2 ATLV01018366 KE525231 KFB43016.1 ADMH02000358 ETN66841.1 JRES01001119 KNC25378.1 AXCN02002096 GDIP01166215 JAJ57187.1 AAAB01008984 APCN01003350 APCN01003351 APCN01003352 CP012523 ALC39302.1 EAA14802.5 DS232030 EDS32530.1 JXUM01029951 JXUM01029952 KQ560868 KXJ80560.1 GDIQ01168500 JAK83225.1 GDIQ01129872 JAL21854.1 GDIQ01053087 JAN41650.1 GDIQ01131119 JAL20607.1 GDIP01046329 JAM57386.1 GDIP01157172 JAJ66230.1 GDIQ01097368 JAL54358.1 GDIQ01085077 JAN09660.1 GDIP01020457 JAM83258.1 GDIQ01160955 JAK90770.1 GDIQ01013644 JAN81093.1 GDIQ01136266 JAL15460.1 EDW88055.1 CH480818 EDW52571.1 CM000361 EDX04675.1 EDV58915.1 X72800 X72801 CH477336 EAT43201.1 CH479180 EDW29126.1 CH916368 EDW02936.1 LRGB01000243 KZS20192.1
KQ971357 KYB26028.1 KYB26029.1 ACPB03001838 GDHC01017635 JAQ00994.1 KK852421 KDR24308.1 PYGN01001181 PSN36852.1 DS235878 EEB19820.1 CH940649 EDW63420.2 KRF81107.1 CH933807 EDW13325.2 APGK01039614 APGK01039615 APGK01039616 KB740972 ENN76628.1 AJVK01002401 AJVK01002402 AJVK01002403 AJVK01002404 GDIQ01052348 JAN42389.1 GDIP01166914 JAJ56488.1 GDIP01167810 JAJ55592.1 GDIP01119023 JAL84691.1 GDIP01086478 JAM17237.1 GDIP01014364 JAM89351.1 GDIP01160124 JAJ63278.1 GDIP01222214 JAJ01188.1 GDIQ01039146 JAN55591.1 GDIP01177777 JAJ45625.1 GDIP01222213 GDIP01222212 JAJ01189.1 GDIP01081672 JAM22043.1 GDIP01189168 JAJ34234.1 GDIP01112873 JAL90841.1 GDIQ01014350 JAN80387.1 GDIQ01039147 JAN55590.1 GDIP01158720 JAJ64682.1 LRGB01002475 KZS07425.1 GDIP01160123 JAJ63279.1 GDIP01160822 JAJ62580.1 GDIP01102876 JAM00839.1 KK855823 PTY23999.1 OUUW01000004 SPP79483.1 CH379060 EDY70153.2 KRT04269.1 CM000157 KRJ97540.1 KRT04268.1 JXJN01021989 JXJN01021990 CM002910 KMY89709.1 CCAG010004953 CH954177 KQS70572.1 AE014134 ABV53670.1 CH902620 EDV30989.2 CH963857 EDW76011.2 ATLV01018366 KE525231 KFB43016.1 ADMH02000358 ETN66841.1 JRES01001119 KNC25378.1 AXCN02002096 GDIP01166215 JAJ57187.1 AAAB01008984 APCN01003350 APCN01003351 APCN01003352 CP012523 ALC39302.1 EAA14802.5 DS232030 EDS32530.1 JXUM01029951 JXUM01029952 KQ560868 KXJ80560.1 GDIQ01168500 JAK83225.1 GDIQ01129872 JAL21854.1 GDIQ01053087 JAN41650.1 GDIQ01131119 JAL20607.1 GDIP01046329 JAM57386.1 GDIP01157172 JAJ66230.1 GDIQ01097368 JAL54358.1 GDIQ01085077 JAN09660.1 GDIP01020457 JAM83258.1 GDIQ01160955 JAK90770.1 GDIQ01013644 JAN81093.1 GDIQ01136266 JAL15460.1 EDW88055.1 CH480818 EDW52571.1 CM000361 EDX04675.1 EDV58915.1 X72800 X72801 CH477336 EAT43201.1 CH479180 EDW29126.1 CH916368 EDW02936.1 LRGB01000243 KZS20192.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007266
UP000015103
UP000027135
+ More
UP000245037 UP000009046 UP000008792 UP000009192 UP000019118 UP000092462 UP000076858 UP000268350 UP000001819 UP000002282 UP000078200 UP000092460 UP000092445 UP000092444 UP000192221 UP000008711 UP000000803 UP000007801 UP000007798 UP000095300 UP000030765 UP000069272 UP000000673 UP000075880 UP000037069 UP000076408 UP000075886 UP000075884 UP000075900 UP000095301 UP000075903 UP000075920 UP000076407 UP000075840 UP000075902 UP000075882 UP000092553 UP000007062 UP000002320 UP000069940 UP000249989 UP000075885 UP000075901 UP000092443 UP000001292 UP000000304 UP000008820 UP000008744 UP000001070
UP000245037 UP000009046 UP000008792 UP000009192 UP000019118 UP000092462 UP000076858 UP000268350 UP000001819 UP000002282 UP000078200 UP000092460 UP000092445 UP000092444 UP000192221 UP000008711 UP000000803 UP000007801 UP000007798 UP000095300 UP000030765 UP000069272 UP000000673 UP000075880 UP000037069 UP000076408 UP000075886 UP000075884 UP000075900 UP000095301 UP000075903 UP000075920 UP000076407 UP000075840 UP000075902 UP000075882 UP000092553 UP000007062 UP000002320 UP000069940 UP000249989 UP000075885 UP000075901 UP000092443 UP000001292 UP000000304 UP000008820 UP000008744 UP000001070
PRIDE
Pfam
Interpro
IPR011009
Kinase-like_dom_sf
+ More
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001828 ANF_lig-bd_rcpt
IPR029787 Nucleotide_cyclase
IPR000719 Prot_kinase_dom
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR018297 A/G_cyclase_CS
IPR001170 ANPR/GUC
IPR011645 HNOB_dom_associated
IPR008266 Tyr_kinase_AS
IPR036291 NAD(P)-bd_dom_sf
IPR006140 D-isomer_DH_NAD-bd
IPR006139 D-isomer_2_OHA_DH_cat_dom
IPR029753 D-isomer_DH_CS
IPR029752 D-isomer_DH_CS1
IPR041470 GCP_N
IPR042241 GCP_C_sf
IPR040457 GCP_C
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR001828 ANF_lig-bd_rcpt
IPR029787 Nucleotide_cyclase
IPR000719 Prot_kinase_dom
IPR001054 A/G_cyclase
IPR028082 Peripla_BP_I
IPR018297 A/G_cyclase_CS
IPR001170 ANPR/GUC
IPR011645 HNOB_dom_associated
IPR008266 Tyr_kinase_AS
IPR036291 NAD(P)-bd_dom_sf
IPR006140 D-isomer_DH_NAD-bd
IPR006139 D-isomer_2_OHA_DH_cat_dom
IPR029753 D-isomer_DH_CS
IPR029752 D-isomer_DH_CS1
IPR041470 GCP_N
IPR042241 GCP_C_sf
IPR040457 GCP_C
Gene 3D
ProteinModelPortal
A0A194QSQ1
A0A194QAM5
A0A2A4JP88
A0A139WDI2
A0A139WDK0
T1HNK9
+ More
A0A146KWY4 A0A067RMP4 A0A2P8XXW2 E0W2G4 B4LQH0 B4KHA8 N6TFV7 A0A1B0D0D2 A0A0N8E865 A0A0P5CQL6 A0A0P5CN85 A0A0N8CLV3 A0A0P5WH20 A0A0P6BHS8 A0A0P5EGZ5 A0A0P4YXA1 A0A0P6G379 A0A0N8A6H0 A0A0P4ZRW1 A0A0P5WUA0 A0A0P5B5W5 A0A0P5UHG3 A0A0P6HWV4 A0A0P6G382 A0A0P5DR45 A0A164Q4I2 A0A0P5DGF2 A0A0P5D5R6 A0A0P5VEM9 A0A2R7WWS8 A0A3B0JFK9 B5DIL1 A0A0R3NYC5 A0A0R1DJU5 A0A0R3NTK6 A0A1A9UXC2 A0A1B0BWX4 A0A1B0A0T9 A0A0J9R1J4 A0A1B0FQA5 A0A1W4W5P4 A0A0Q5WB16 A8DYZ3 B3MMW4 B4MWE1 A0A1I8PD95 A0A084VYH2 A0A182F1Z3 W5JRT6 A0A182ILP4 A0A0L0BZE8 A0A182Y970 A0A182Q3F5 A0A182NP43 A0A182R498 A0A0P5CS31 A0A1I8NG04 A0A182VBW3 A0A182VU62 A0A1S4H063 A0A182XDI7 A0A2C9GRZ6 A0A182U4U3 A0A182KNH9 A0A0M4E8L4 Q7PWU5 B0WPU2 A0A1S4FAA0 A0A182H963 A0A0P5LTI7 A0A0P5QU44 A0A0P6FWN6 A0A0P5PAZ8 A0A0P5ZAY1 A0A0P5DI19 A0A0P5RTM9 A0A0P6DLI3 A0A0P6B8G8 A0A0N8BNB3 A0A0N8EM03 A0A0P5NXS8 A0A182P9Q0 A0A182SNN7 A0A1A9Y362 B4P180 B4HX26 B4QAC0 B3N4K3 Q07553 Q17AF9 B4G9U6 B4JBD1 A0A162R3A0
A0A146KWY4 A0A067RMP4 A0A2P8XXW2 E0W2G4 B4LQH0 B4KHA8 N6TFV7 A0A1B0D0D2 A0A0N8E865 A0A0P5CQL6 A0A0P5CN85 A0A0N8CLV3 A0A0P5WH20 A0A0P6BHS8 A0A0P5EGZ5 A0A0P4YXA1 A0A0P6G379 A0A0N8A6H0 A0A0P4ZRW1 A0A0P5WUA0 A0A0P5B5W5 A0A0P5UHG3 A0A0P6HWV4 A0A0P6G382 A0A0P5DR45 A0A164Q4I2 A0A0P5DGF2 A0A0P5D5R6 A0A0P5VEM9 A0A2R7WWS8 A0A3B0JFK9 B5DIL1 A0A0R3NYC5 A0A0R1DJU5 A0A0R3NTK6 A0A1A9UXC2 A0A1B0BWX4 A0A1B0A0T9 A0A0J9R1J4 A0A1B0FQA5 A0A1W4W5P4 A0A0Q5WB16 A8DYZ3 B3MMW4 B4MWE1 A0A1I8PD95 A0A084VYH2 A0A182F1Z3 W5JRT6 A0A182ILP4 A0A0L0BZE8 A0A182Y970 A0A182Q3F5 A0A182NP43 A0A182R498 A0A0P5CS31 A0A1I8NG04 A0A182VBW3 A0A182VU62 A0A1S4H063 A0A182XDI7 A0A2C9GRZ6 A0A182U4U3 A0A182KNH9 A0A0M4E8L4 Q7PWU5 B0WPU2 A0A1S4FAA0 A0A182H963 A0A0P5LTI7 A0A0P5QU44 A0A0P6FWN6 A0A0P5PAZ8 A0A0P5ZAY1 A0A0P5DI19 A0A0P5RTM9 A0A0P6DLI3 A0A0P6B8G8 A0A0N8BNB3 A0A0N8EM03 A0A0P5NXS8 A0A182P9Q0 A0A182SNN7 A0A1A9Y362 B4P180 B4HX26 B4QAC0 B3N4K3 Q07553 Q17AF9 B4G9U6 B4JBD1 A0A162R3A0
Ontologies
KEGG
GO
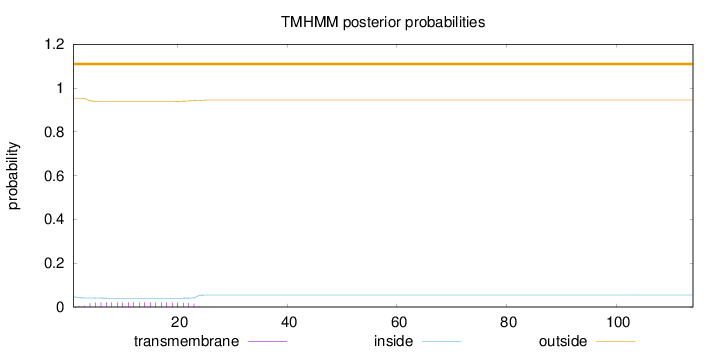
Topology
Subcellular location
Membrane
Length:
114
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.42363
Exp number, first 60 AAs:
0.42345
Total prob of N-in:
0.04705
outside
1 - 114
Population Genetic Test Statistics
Pi
191.050775
Theta
152.786834
Tajima's D
1.695688
CLR
0.44716
CSRT
0.833858307084646
Interpretation
Uncertain
