Gene
KWMTBOMO10073
Pre Gene Modal
BGIBMGA005673
Annotation
acidic_lipase_[Helicoverpa_armigera]
Full name
Lipase
Location in the cell
PlasmaMembrane Reliability : 2.896
Sequence
CDS
ATGATGCTGCTAATTTTATTTCTGTGCATCGGAGCTGTTGCAGGGAGGAGATCACCGCATGCAGAGTACATTGAGGAGCTGGAACAGAGTCTGGTTGGTACCCCAAGCGATGTGCTGGAAGATGCTAAACTCGACGTGCCTGGTCTTCTAATCAAATACGGATATCCCTCCGAGATCCACAGTGTAGAAGCAGAAGATGGTTACATACTGGAAATGCACCGGATTCCTCACGGTAGGAACGAAGGGAGCAGACCTAAACAGAATAGACCGGTGGTCTTCCTGATGCATGGTCTTTTATCGTCTTCAGCTGACTTCTTGCTACTAGGGCCGAGTACTGCTTTAGGTTACATCCTATCCGACGCCGGCTATGATGTCTGGTTGGGTAACGCCCGCGGTAACTTCTACTCTCGCAAGCACAGACGTCACAATCCGGACAGCCTCATCACTCAGAGATTTTGGAAGTTCAGCTGGGACGAAATAGGCAATTACGACCTACCGGCCATGATAGATTACGTTTTAGAGGCCACCGGGCAACAGAAAGTTCACTATGTCGGCCATTCCCAAGGCGGAACCACGTTTTTGGTGTTAAATTCGTTAAGACCGGAATATAATGAGAAGTTCATTTCCTTCCAAGGGTTGGCACCTGCTTCCTTCTTCACATACAATGAACATTCAACTTTCAACGCTCTGGCGCCGTATGAAAATGTTCTTGAGAGTGCAGCCTTTGCTATGGGTGTGGGTGAAATCTTTGGTAACAGGGAATTCATGACCTGGTTCGCCACCAACCATTGCATCGAGGACTCGATTTTGTTTCCCTTGTGCACCAACACGCTACCCGGTATTACTTCCTCTGAATATTTTAACTCGACTTTGTTGCCACTGATCCTTGGCCACACTCCAGCCGGTGCATCCATCCGGCAAGTAGCCCACTACGGTCAGATCATTCGTTTTGACGCCTTCCGCCGCTACGACCACGATCCGATCAGAAACCTCATCAACTACGGCCAATTGCAGCCACCGAACTATGACCTCAGTAAAATCACGGTCCCAGCTTATCTCCACTACGGCTTAAAGGACAGGCAAGTCGACTACAGGGATCTTTACCTTTTGGCCAGGAAATTGCCAAACACAATAGGCGTTTACAAAGTGGCGCGTGATAGCTTCAATCACTTCGATTTTCTTTGGGCCTCTGATGTCAAAGAAATGTTTTATGATAGACTATTGGAGCACTTAAAACTTGCGGAAGCGCAATTTTTGTAA
Protein
MMLLILFLCIGAVAGRRSPHAEYIEELEQSLVGTPSDVLEDAKLDVPGLLIKYGYPSEIHSVEAEDGYILEMHRIPHGRNEGSRPKQNRPVVFLMHGLLSSSADFLLLGPSTALGYILSDAGYDVWLGNARGNFYSRKHRRHNPDSLITQRFWKFSWDEIGNYDLPAMIDYVLEATGQQKVHYVGHSQGGTTFLVLNSLRPEYNEKFISFQGLAPASFFTYNEHSTFNALAPYENVLESAAFAMGVGEIFGNREFMTWFATNHCIEDSILFPLCTNTLPGITSSEYFNSTLLPLILGHTPAGASIRQVAHYGQIIRFDAFRRYDHDPIRNLINYGQLQPPNYDLSKITVPAYLHYGLKDRQVDYRDLYLLARKLPNTIGVYKVARDSFNHFDFLWASDVKEMFYDRLLEHLKLAEAQFL
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
H9J831
I3QC09
A0A2A4JD47
A0A2H1X102
I1SR89
A0A0L7L5L9
+ More
A0A3S2NF67 F5GTG4 H9JQH1 A0A2A4K6W6 A0A2A4K660 A0A2A4K5R5 A0A194QNQ2 A0A2A4K5V7 A0A3S2NJM8 H9JM87 A0A2H1WZN4 I3QC10 A0A2H1WEG8 A0A194PVH3 H9JM86 A0A194QNR0 A0A194Q1W6 A0A194PXA6 A0A194QQB7 A0A194PWK6 A0A194QUC1 A0A3S2LGI4 A0A212FG21 A0A194PVN8 A0A2H1VT83 A0A1E1W7Q1 A0A2A4K595 A0A1E1VY19 A0A212F4Q9 A0A2A4IVE2 A0A194R2Y3 A0A194PL00 A0A084W6W2 A0A212FB51 A0A182JMM3 A0A2H1X0E5 A0A1W4VT50 A0A3S2P1G5 B4Q9R3 Q29KR3 B3N558 A0A067RCF7 B4HWT2 Q9VKS5 B4GSX7 A0A0A1XAM0 A0A3B0JN49 B4P0S2 B4JE79 B4MWQ5 B4KKQ8 B3MJL9 A0A0K8VTT0 A0A0L0BXN7 A0A0K8VS63 B4LUJ8 A0A0M4E6C0 A0A034WLL0 Q16F27 Q16MD1 A0A1S4FVZ7 A0A1I8PCV0 W5JF51 A0A0C9RRQ5 A0A182H3N2 A0A1B0A014 A0A151X458 A0A1B0B8Z4 A0A1B0G0B6 U5ERJ2 A0A182FR70 A0A1L8DQ21 A0A1L8DYQ8 W8ADN0 B0W6G8 A0A1I8NAV5 A0A1A9UTK2 A0A182RCW3 A0A195CQI8 A0A1L8DQ28 E2A1A9 A0A1L8DPS2 A0A182H3N6 A0A182GFS7 A0A026W0L2 A0A139WJS1 E9I8R3 A0A182J3W9 A0A2P8XRN9 K7J8S8 D2A0X3 Q16MD4 A0A1L8DPY1 Q16F26
A0A3S2NF67 F5GTG4 H9JQH1 A0A2A4K6W6 A0A2A4K660 A0A2A4K5R5 A0A194QNQ2 A0A2A4K5V7 A0A3S2NJM8 H9JM87 A0A2H1WZN4 I3QC10 A0A2H1WEG8 A0A194PVH3 H9JM86 A0A194QNR0 A0A194Q1W6 A0A194PXA6 A0A194QQB7 A0A194PWK6 A0A194QUC1 A0A3S2LGI4 A0A212FG21 A0A194PVN8 A0A2H1VT83 A0A1E1W7Q1 A0A2A4K595 A0A1E1VY19 A0A212F4Q9 A0A2A4IVE2 A0A194R2Y3 A0A194PL00 A0A084W6W2 A0A212FB51 A0A182JMM3 A0A2H1X0E5 A0A1W4VT50 A0A3S2P1G5 B4Q9R3 Q29KR3 B3N558 A0A067RCF7 B4HWT2 Q9VKS5 B4GSX7 A0A0A1XAM0 A0A3B0JN49 B4P0S2 B4JE79 B4MWQ5 B4KKQ8 B3MJL9 A0A0K8VTT0 A0A0L0BXN7 A0A0K8VS63 B4LUJ8 A0A0M4E6C0 A0A034WLL0 Q16F27 Q16MD1 A0A1S4FVZ7 A0A1I8PCV0 W5JF51 A0A0C9RRQ5 A0A182H3N2 A0A1B0A014 A0A151X458 A0A1B0B8Z4 A0A1B0G0B6 U5ERJ2 A0A182FR70 A0A1L8DQ21 A0A1L8DYQ8 W8ADN0 B0W6G8 A0A1I8NAV5 A0A1A9UTK2 A0A182RCW3 A0A195CQI8 A0A1L8DQ28 E2A1A9 A0A1L8DPS2 A0A182H3N6 A0A182GFS7 A0A026W0L2 A0A139WJS1 E9I8R3 A0A182J3W9 A0A2P8XRN9 K7J8S8 D2A0X3 Q16MD4 A0A1L8DPY1 Q16F26
Pubmed
19121390
26227816
21910995
26354079
22118469
24438588
+ More
17994087 22936249 15632085 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17550304 26108605 18057021 25348373 17510324 20920257 23761445 26483478 24495485 25315136 20798317 24508170 30249741 18362917 19820115 21282665 29403074 20075255
17994087 22936249 15632085 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 17550304 26108605 18057021 25348373 17510324 20920257 23761445 26483478 24495485 25315136 20798317 24508170 30249741 18362917 19820115 21282665 29403074 20075255
EMBL
BABH01020326
JF999958
AFI64312.1
NWSH01001918
PCG69686.1
ODYU01012554
+ More
SOQ58947.1 JF303742 ADZ54058.1 JTDY01002861 KOB70584.1 RSAL01000145 RVE45945.1 JI484237 AEB26288.1 BABH01011980 NWSH01000114 PCG79412.1 PCG79408.1 PCG79409.1 KQ461190 KPJ07153.1 PCG79406.1 RSAL01000448 RVE41683.1 BABH01033349 BABH01033350 BABH01033351 BABH01033352 ODYU01012265 SOQ58477.1 JF999959 AFI64313.1 ODYU01008119 SOQ51451.1 KQ459590 KPI97391.1 BABH01033355 KPJ07152.1 KPI97395.1 KPI97394.1 KPJ07150.1 KPI97393.1 KPJ07151.1 RSAL01000137 RVE46199.1 AGBW02008724 OWR52685.1 KPI97392.1 ODYU01004000 SOQ43434.1 GDQN01008173 JAT82881.1 PCG79407.1 GDQN01011447 JAT79607.1 AGBW02010324 OWR48717.1 NWSH01005801 PCG63937.1 KQ460855 KPJ11879.1 KQ459601 KPI94007.1 ATLV01020990 KE525311 KFB45956.1 AGBW02009374 OWR50972.1 ODYU01012460 SOQ58781.1 RSAL01000004 RVE54656.1 CM000361 CM002910 EDX04580.1 KMY89593.1 CH379061 EAL33110.2 CH954177 EDV59003.1 KK852794 KDR16428.1 CH480818 EDW52477.1 AE014134 BT003518 AAF52985.2 AAO39522.1 CH479189 EDW25486.1 GBXI01006321 JAD07971.1 OUUW01000006 SPP81772.1 CM000157 EDW87967.1 CH916368 EDW03599.1 CH963857 EDW76544.2 CH933807 EDW12722.1 CH902620 EDV32387.1 GDHF01010028 GDHF01008682 JAI42286.1 JAI43632.1 JRES01001173 KNC24792.1 GDHF01010621 JAI41693.1 CH940649 EDW64184.1 KRF81481.1 CP012523 ALC38102.1 GAKP01004299 JAC54653.1 CH478517 EAT32843.1 CH477866 EAT35487.1 ADMH02001587 ETN61958.1 GBYB01010056 JAG79823.1 JXUM01107700 KQ565165 KXJ71265.1 KQ982557 KYQ55059.1 JXJN01010151 JXJN01010152 JXJN01010153 JXJN01010154 CCAG010008921 GANO01002781 JAB57090.1 GFDF01005639 JAV08445.1 GFDF01002510 JAV11574.1 GAMC01020340 JAB86215.1 DS231848 EDS36642.1 KQ977481 KYN02379.1 GFDF01005644 JAV08440.1 GL435739 EFN72780.1 GFDF01005645 JAV08439.1 JXUM01107701 KXJ71269.1 JXUM01060248 KQ562094 KXJ76704.1 KK107599 QOIP01000012 EZA48594.1 RLU15932.1 KQ971338 KYB28091.1 GL761642 EFZ23043.1 PYGN01001466 PSN34671.1 EFA01616.1 EAT35484.1 GFDF01005647 JAV08437.1 EAT32844.1
SOQ58947.1 JF303742 ADZ54058.1 JTDY01002861 KOB70584.1 RSAL01000145 RVE45945.1 JI484237 AEB26288.1 BABH01011980 NWSH01000114 PCG79412.1 PCG79408.1 PCG79409.1 KQ461190 KPJ07153.1 PCG79406.1 RSAL01000448 RVE41683.1 BABH01033349 BABH01033350 BABH01033351 BABH01033352 ODYU01012265 SOQ58477.1 JF999959 AFI64313.1 ODYU01008119 SOQ51451.1 KQ459590 KPI97391.1 BABH01033355 KPJ07152.1 KPI97395.1 KPI97394.1 KPJ07150.1 KPI97393.1 KPJ07151.1 RSAL01000137 RVE46199.1 AGBW02008724 OWR52685.1 KPI97392.1 ODYU01004000 SOQ43434.1 GDQN01008173 JAT82881.1 PCG79407.1 GDQN01011447 JAT79607.1 AGBW02010324 OWR48717.1 NWSH01005801 PCG63937.1 KQ460855 KPJ11879.1 KQ459601 KPI94007.1 ATLV01020990 KE525311 KFB45956.1 AGBW02009374 OWR50972.1 ODYU01012460 SOQ58781.1 RSAL01000004 RVE54656.1 CM000361 CM002910 EDX04580.1 KMY89593.1 CH379061 EAL33110.2 CH954177 EDV59003.1 KK852794 KDR16428.1 CH480818 EDW52477.1 AE014134 BT003518 AAF52985.2 AAO39522.1 CH479189 EDW25486.1 GBXI01006321 JAD07971.1 OUUW01000006 SPP81772.1 CM000157 EDW87967.1 CH916368 EDW03599.1 CH963857 EDW76544.2 CH933807 EDW12722.1 CH902620 EDV32387.1 GDHF01010028 GDHF01008682 JAI42286.1 JAI43632.1 JRES01001173 KNC24792.1 GDHF01010621 JAI41693.1 CH940649 EDW64184.1 KRF81481.1 CP012523 ALC38102.1 GAKP01004299 JAC54653.1 CH478517 EAT32843.1 CH477866 EAT35487.1 ADMH02001587 ETN61958.1 GBYB01010056 JAG79823.1 JXUM01107700 KQ565165 KXJ71265.1 KQ982557 KYQ55059.1 JXJN01010151 JXJN01010152 JXJN01010153 JXJN01010154 CCAG010008921 GANO01002781 JAB57090.1 GFDF01005639 JAV08445.1 GFDF01002510 JAV11574.1 GAMC01020340 JAB86215.1 DS231848 EDS36642.1 KQ977481 KYN02379.1 GFDF01005644 JAV08440.1 GL435739 EFN72780.1 GFDF01005645 JAV08439.1 JXUM01107701 KXJ71269.1 JXUM01060248 KQ562094 KXJ76704.1 KK107599 QOIP01000012 EZA48594.1 RLU15932.1 KQ971338 KYB28091.1 GL761642 EFZ23043.1 PYGN01001466 PSN34671.1 EFA01616.1 EAT35484.1 GFDF01005647 JAV08437.1 EAT32844.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000283053
UP000053240
UP000053268
+ More
UP000007151 UP000030765 UP000075880 UP000192221 UP000000304 UP000001819 UP000008711 UP000027135 UP000001292 UP000000803 UP000008744 UP000268350 UP000002282 UP000001070 UP000007798 UP000009192 UP000007801 UP000037069 UP000008792 UP000092553 UP000008820 UP000095300 UP000000673 UP000069940 UP000249989 UP000092445 UP000075809 UP000092460 UP000092444 UP000069272 UP000002320 UP000095301 UP000078200 UP000075900 UP000078542 UP000000311 UP000053097 UP000279307 UP000007266 UP000245037 UP000002358
UP000007151 UP000030765 UP000075880 UP000192221 UP000000304 UP000001819 UP000008711 UP000027135 UP000001292 UP000000803 UP000008744 UP000268350 UP000002282 UP000001070 UP000007798 UP000009192 UP000007801 UP000037069 UP000008792 UP000092553 UP000008820 UP000095300 UP000000673 UP000069940 UP000249989 UP000092445 UP000075809 UP000092460 UP000092444 UP000069272 UP000002320 UP000095301 UP000078200 UP000075900 UP000078542 UP000000311 UP000053097 UP000279307 UP000007266 UP000245037 UP000002358
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9J831
I3QC09
A0A2A4JD47
A0A2H1X102
I1SR89
A0A0L7L5L9
+ More
A0A3S2NF67 F5GTG4 H9JQH1 A0A2A4K6W6 A0A2A4K660 A0A2A4K5R5 A0A194QNQ2 A0A2A4K5V7 A0A3S2NJM8 H9JM87 A0A2H1WZN4 I3QC10 A0A2H1WEG8 A0A194PVH3 H9JM86 A0A194QNR0 A0A194Q1W6 A0A194PXA6 A0A194QQB7 A0A194PWK6 A0A194QUC1 A0A3S2LGI4 A0A212FG21 A0A194PVN8 A0A2H1VT83 A0A1E1W7Q1 A0A2A4K595 A0A1E1VY19 A0A212F4Q9 A0A2A4IVE2 A0A194R2Y3 A0A194PL00 A0A084W6W2 A0A212FB51 A0A182JMM3 A0A2H1X0E5 A0A1W4VT50 A0A3S2P1G5 B4Q9R3 Q29KR3 B3N558 A0A067RCF7 B4HWT2 Q9VKS5 B4GSX7 A0A0A1XAM0 A0A3B0JN49 B4P0S2 B4JE79 B4MWQ5 B4KKQ8 B3MJL9 A0A0K8VTT0 A0A0L0BXN7 A0A0K8VS63 B4LUJ8 A0A0M4E6C0 A0A034WLL0 Q16F27 Q16MD1 A0A1S4FVZ7 A0A1I8PCV0 W5JF51 A0A0C9RRQ5 A0A182H3N2 A0A1B0A014 A0A151X458 A0A1B0B8Z4 A0A1B0G0B6 U5ERJ2 A0A182FR70 A0A1L8DQ21 A0A1L8DYQ8 W8ADN0 B0W6G8 A0A1I8NAV5 A0A1A9UTK2 A0A182RCW3 A0A195CQI8 A0A1L8DQ28 E2A1A9 A0A1L8DPS2 A0A182H3N6 A0A182GFS7 A0A026W0L2 A0A139WJS1 E9I8R3 A0A182J3W9 A0A2P8XRN9 K7J8S8 D2A0X3 Q16MD4 A0A1L8DPY1 Q16F26
A0A3S2NF67 F5GTG4 H9JQH1 A0A2A4K6W6 A0A2A4K660 A0A2A4K5R5 A0A194QNQ2 A0A2A4K5V7 A0A3S2NJM8 H9JM87 A0A2H1WZN4 I3QC10 A0A2H1WEG8 A0A194PVH3 H9JM86 A0A194QNR0 A0A194Q1W6 A0A194PXA6 A0A194QQB7 A0A194PWK6 A0A194QUC1 A0A3S2LGI4 A0A212FG21 A0A194PVN8 A0A2H1VT83 A0A1E1W7Q1 A0A2A4K595 A0A1E1VY19 A0A212F4Q9 A0A2A4IVE2 A0A194R2Y3 A0A194PL00 A0A084W6W2 A0A212FB51 A0A182JMM3 A0A2H1X0E5 A0A1W4VT50 A0A3S2P1G5 B4Q9R3 Q29KR3 B3N558 A0A067RCF7 B4HWT2 Q9VKS5 B4GSX7 A0A0A1XAM0 A0A3B0JN49 B4P0S2 B4JE79 B4MWQ5 B4KKQ8 B3MJL9 A0A0K8VTT0 A0A0L0BXN7 A0A0K8VS63 B4LUJ8 A0A0M4E6C0 A0A034WLL0 Q16F27 Q16MD1 A0A1S4FVZ7 A0A1I8PCV0 W5JF51 A0A0C9RRQ5 A0A182H3N2 A0A1B0A014 A0A151X458 A0A1B0B8Z4 A0A1B0G0B6 U5ERJ2 A0A182FR70 A0A1L8DQ21 A0A1L8DYQ8 W8ADN0 B0W6G8 A0A1I8NAV5 A0A1A9UTK2 A0A182RCW3 A0A195CQI8 A0A1L8DQ28 E2A1A9 A0A1L8DPS2 A0A182H3N6 A0A182GFS7 A0A026W0L2 A0A139WJS1 E9I8R3 A0A182J3W9 A0A2P8XRN9 K7J8S8 D2A0X3 Q16MD4 A0A1L8DPY1 Q16F26
PDB
1HLG
E-value=5.63877e-68,
Score=654
Ontologies
GO
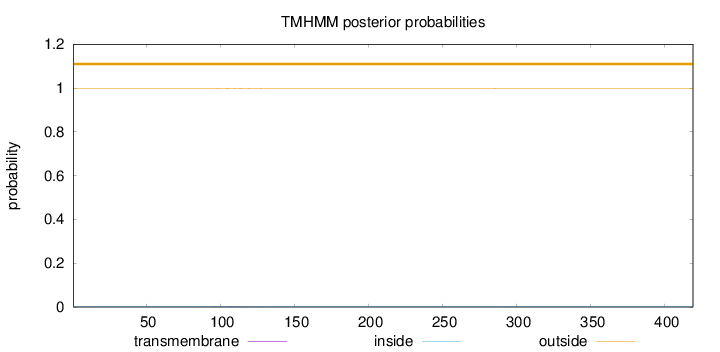
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.980664
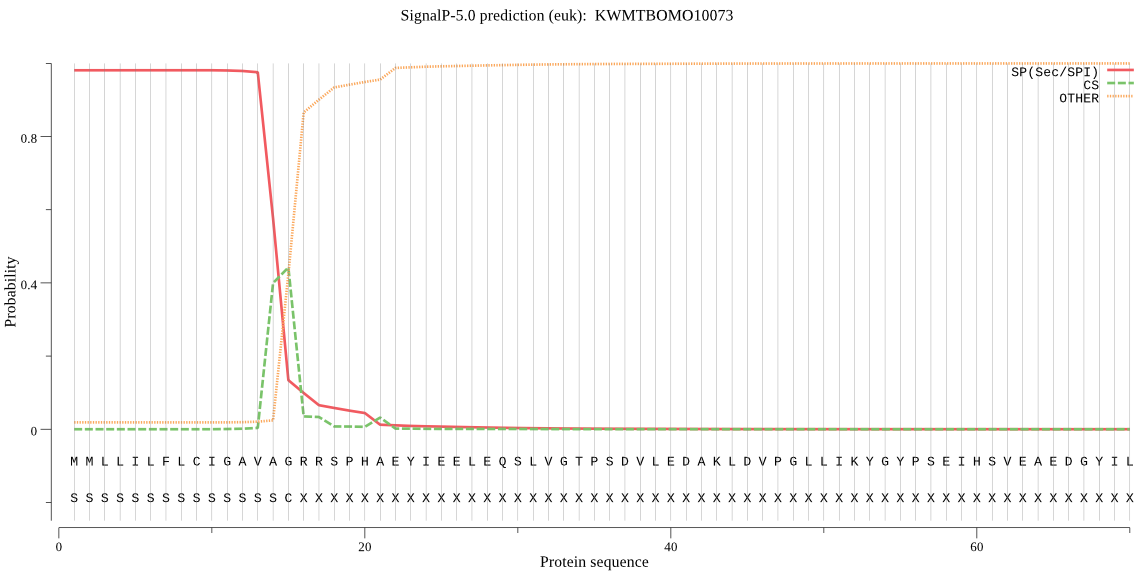
Length:
419
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0719499999999999
Exp number, first 60 AAs:
0.00233
Total prob of N-in:
0.00237
outside
1 - 419
Population Genetic Test Statistics
Pi
269.234299
Theta
152.973008
Tajima's D
1.754875
CLR
0.365952
CSRT
0.83850807459627
Interpretation
Uncertain
