Gene
KWMTBOMO10066
Pre Gene Modal
BGIBMGA005539
Annotation
putative_chitinase_3_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.436
Sequence
CDS
ATGGCGTATTCATATCGGCCACCGATATTGCTTCGATGGATGCTCACCGTGGTTCTGGTCATAGCTGTCATCGCTCCCACGTCAGACTCAACAACTGTCCGCAGAAGACTCCGGAAGCCATCAAAGGTGTCCACATCGGTTTCGTCCAGCGTCTCTAGATCCACAGATCAAGTTCTCTCAGCTAGCGTGAACAGACCAAAGATTCGTGGTCGTCCTTCAATAGCCAGTAGAAAATCATCCGCGGCTCTGGACAACTCCGTCACCACCGAACATCACAAAGATAAGGATGGATACAAGATCGTCTGCTATTACACCAACTGGTCCCAATACCGGACGAAGGTCGGGAAGTTCGTTCCAGAGGACATTCAGCCCGATCTCTGCACCCACATCATCTTCGCTTTCGGCTGGCTGAAGAAAGGAAAACTGAGCAGCTTCGAATCGAACGATGAGACGAAAGACGGGAAGACAGGACTGTACGACAAGATTAATGGGCTGAAGAAAGGGAATCCTAAACTAAAGACTCTATTGGCCATTGGCGGTTGGTCTTTCGGGACTCAGAAGTTCAAGGACATGGTGGCTACCAGATACGCCCGGCAGACGTTCATCTACTCCGCGATTCCTTACTTGAGGGACAGGAACTTTGACGGGCTCGACATCGACTGGGAATACCCCAAGGGTGGAGACGACAAGAAAAACTACGTGCTCCTGTTGAAAGAGCTCCGTGAGGCCTTCGAAGCGGAGGCTCAAGAAGTGAAGAAACCTCGTTTGCTTCTTACTGCAGCTGTGCCAGTTGGACCTGACAACATCAAGAGCGGATACGACGTTCCTGCTGTAGCCAGCTATCTGGACTTCATCAACCTTATGGCCTACGACTTCCACGGCAAATGGGAGAGGGAGACCGGCCACAACGCCCCGCTGTACGCTCCATCGACAGACTCGGAGTGGAGGAAGCAACTATCAGTAGACCACGCAGCACATCTCTGGGTGAAACTTGGAGCCCCGAAGGACAAACTTATTATTGGTATGCCAACATATGGACGTACATTCACGCTCTCCAACCCTAATAACTTCAAAGTGAACGCGCCGGCCAGTGGTGGAGGAAAAGCGGGAGAGTACACCAAGGAAAGCGGTTTCCTCGCTTACTATGAGGTGTGCGAAATGCTAAGGAACGGGGGTGCCTACGTCTGGGACGATGAAATGAAAGTCCCTTACTGCGTCCACGGGGACCAGTGGGTCGGATTCGACGATGAGAAATCAATCAGGAACAAAATGAGGTGGATCAAAGACAATGGCTTCGGAGGAGCAATGGTGTGGACCGTTGATATGGACGACTTCTCCGGCAACGTCTGCGGCGGCGATGTAAAGTATCCATTGATCGGTGCAATGAGGGAAGAACTCCGAGGTATATCTCGAGGTAAAGACGCCAAAGATGTCGACTGGGCCACGGTTGCTTCCAGCATAGTGATAGAAGAAACTGAGAAACCAGAACCGATACAGGTTCCATTGTCCGAAGTCCTCAAGCGCATCAAGAAGCCTGGTAAAAACGTCATCATAAAGACCAATGGAGCGGCACTTTCAGATAAAAACAAACGCGAGCCCCAAGTACTCTGTTATCTCACATCCTGGTCAGCCAAACGTCCAAGCGCAGGCCGCTTCACACCAGAAAACGTTGACCCCAAACTCTGTACCCACATCATATACGCTTTCGCAACCCTCAAAGACCACAAGCTAGCTGAAGCTGACGACAAAGACGCTGATATGTACGATAAAGTCGTTGCGTTGAGAGAGAAGAACCCCAATTTGAAGATATTGCTCGCCATCGGAGGCTGGGCCTTCGGATCTACTCCGTTCAAAGAATTGACTTCGAACGTGTTCCGCATGAACCAGTTCGTGTATGAAGCCATCGAGTTCCTCCGCGACTATCAGTTCAATGGCTTGGACGTGGATTGGGAATATCCTAGAGGAGCAGATGATCGTGCAGCATTCGTATCTCTGCTCAAGGAATTGCGTCTGGCGTTCGAGGGTGAAGCCAAGACTTCAGGTCAGCCCCGGCTCCTGCTCACTGCTGCTGTACCGGCGTCGTTTGAGGCCATCGCCGCCGGATATGATGTACCGGAAATATCGAAATATTTGGACTTCATAAACGTGATGACGTATGACTTCCACGGCCAGTGGGAGCGCCAGGTCGGCCACAACAGTCCGCTGTTCCCTCTAGAAAGCGCCACGAGCTACCAGAAGAAACTAACTGTGGACTACTCCGCCCGCGAGTGGGTCCGCCAGGGCGCCCCCAAAGAGAAGCTGATGATCGGCATGCCCACGTACGGAAGGTCCTTCACGCTTATCAACGAAACACAGTACGACATCGGCGCTCCGGCTTCAGAGGGCGGAGAAGCCGGACGTTTCACCAACGAAGCCGGCTTCATGTCCTACTACGAAATCTGCGAGTTCCTACGAGAGGACAACACGACGCTCGTCTGGGACAACGAGCAGATGGTGCCATTCGCTTACCGAGGCGACCAGTGGGTCGGGTTCGACGACGAGAGATCGCTTAAAACAAAAATGGCCTGGCTGAAGGAAGAAGGGTTCGGCGGGATCATGGTCTGGTCTGTCGACATGGACGACTTCCGCGGCTCCTGTGGAACTGGCAAGTTCCCGCTCATCACGACCATGAAGCAGGAACTGTCCGACTACAAGGTCAAGCTGGAATACGACGGGCCCTACGAGACGGTGCTCACCAGCGGACAGTACACCACTAAGGACCCCACTGAGGTGACGTGCGAGGAGGAGGACGGCCACATCTCGTACCACAAGGACCAGGCGGACTGCACCATGTACTACATGTGCGAGGGCGAGCGGAAGCACCACATGCCGTGCCCCTCCAACCTCGTGTTCAACCCCAACGAGAACGTCTGCGATTGGCCGGAGAACGTGGAGGGATGCGCGCACCACACGCAGGCGCCGCCCGCCAGACGATAG
Protein
MAYSYRPPILLRWMLTVVLVIAVIAPTSDSTTVRRRLRKPSKVSTSVSSSVSRSTDQVLSASVNRPKIRGRPSIASRKSSAALDNSVTTEHHKDKDGYKIVCYYTNWSQYRTKVGKFVPEDIQPDLCTHIIFAFGWLKKGKLSSFESNDETKDGKTGLYDKINGLKKGNPKLKTLLAIGGWSFGTQKFKDMVATRYARQTFIYSAIPYLRDRNFDGLDIDWEYPKGGDDKKNYVLLLKELREAFEAEAQEVKKPRLLLTAAVPVGPDNIKSGYDVPAVASYLDFINLMAYDFHGKWERETGHNAPLYAPSTDSEWRKQLSVDHAAHLWVKLGAPKDKLIIGMPTYGRTFTLSNPNNFKVNAPASGGGKAGEYTKESGFLAYYEVCEMLRNGGAYVWDDEMKVPYCVHGDQWVGFDDEKSIRNKMRWIKDNGFGGAMVWTVDMDDFSGNVCGGDVKYPLIGAMREELRGISRGKDAKDVDWATVASSIVIEETEKPEPIQVPLSEVLKRIKKPGKNVIIKTNGAALSDKNKREPQVLCYLTSWSAKRPSAGRFTPENVDPKLCTHIIYAFATLKDHKLAEADDKDADMYDKVVALREKNPNLKILLAIGGWAFGSTPFKELTSNVFRMNQFVYEAIEFLRDYQFNGLDVDWEYPRGADDRAAFVSLLKELRLAFEGEAKTSGQPRLLLTAAVPASFEAIAAGYDVPEISKYLDFINVMTYDFHGQWERQVGHNSPLFPLESATSYQKKLTVDYSAREWVRQGAPKEKLMIGMPTYGRSFTLINETQYDIGAPASEGGEAGRFTNEAGFMSYYEICEFLREDNTTLVWDNEQMVPFAYRGDQWVGFDDERSLKTKMAWLKEEGFGGIMVWSVDMDDFRGSCGTGKFPLITTMKQELSDYKVKLEYDGPYETVLTSGQYTTKDPTEVTCEEEDGHISYHKDQADCTMYYMCEGERKHHMPCPSNLVFNPNENVCDWPENVEGCAHHTQAPPARR
Summary
Similarity
Belongs to the glycosyl hydrolase 18 family.
Uniprot
U5LUV7
A0A194QAT6
I1VX14
A0A221LFZ8
A0A2H4Z0A2
I6QMM8
+ More
A0A2A4J3B9 A0A194QTM7 A0A2J7QAA7 D2A576 Q0Z942 A0A1B6CMH7 A0A067RC35 A0A0C5CUE1 A0A182WWE3 A0A182L3S0 Q7QIJ1 E0VH87 A0A084VDZ0 A0A182N9E9 A0A182Y814 A0A182I6M1 A0A182VKK7 A0A182LZT7 A0A182TW87 A0A182KCD4 A0A182WBE5 W5JDL8 A0A182IMA2 A0A182RJG7 A0A182QSR0 A0A2S2Q2H3 A0A182FGQ4 A0A2H8TNI6 B0X022 Q17BM9 A0A1Q3FMV9 A0A1Q3FMW1 J9K0Y5 A0A2H4Z2U0 A0A2K8JRQ5 A0A0A9WC72 E2B3W5 A0A195F5G8 A0A2I6Q763 A0A151X836 F4WXH8 A0A195DB08 A0A1B0D2K6 A0A195AY20 E2AH26 A0A195C1V2 A0A158NS63 A0A1V0QBC4 A0A154P9H9 A0A336MKJ0 T1IFA7 A0A0C9RP99 A0A2A3ENV6 A0A088AAP6 A0A1J1J5Q0 A0A1W4W1Q5 A0A0K8WKW5 E9JBY3 A0A0M9A9W5 A0A1A9X2U9 A0A1I8N4R4 A0A0J7KXG8 A0A2Z2Z004 A0A0L0CEG1 A0A1I8PQ66 K7IWS2 A0A0L7QRF8 B4N6I7 B3M7E5 B4KWS7 B3NF27 Q9VZV2 B4QP18 A0A0A1X9V2 Q29E30 B4PCV6 B4PCV5 A0A3B0J5V4 B4LGX0 U4U9T5 N6U4J9 A0A1B0AD34 B4HTJ6 A0A1A9VSK3 A0A1A9Y2B6 A0A1B0B9H1 A0A0M5IYA4 A0A2P8XG74 A0A069DWM4 A0A182PIM9 A0A182H9P8 A0A212FNA5 B4H407 A0A2S2NVJ0
A0A2A4J3B9 A0A194QTM7 A0A2J7QAA7 D2A576 Q0Z942 A0A1B6CMH7 A0A067RC35 A0A0C5CUE1 A0A182WWE3 A0A182L3S0 Q7QIJ1 E0VH87 A0A084VDZ0 A0A182N9E9 A0A182Y814 A0A182I6M1 A0A182VKK7 A0A182LZT7 A0A182TW87 A0A182KCD4 A0A182WBE5 W5JDL8 A0A182IMA2 A0A182RJG7 A0A182QSR0 A0A2S2Q2H3 A0A182FGQ4 A0A2H8TNI6 B0X022 Q17BM9 A0A1Q3FMV9 A0A1Q3FMW1 J9K0Y5 A0A2H4Z2U0 A0A2K8JRQ5 A0A0A9WC72 E2B3W5 A0A195F5G8 A0A2I6Q763 A0A151X836 F4WXH8 A0A195DB08 A0A1B0D2K6 A0A195AY20 E2AH26 A0A195C1V2 A0A158NS63 A0A1V0QBC4 A0A154P9H9 A0A336MKJ0 T1IFA7 A0A0C9RP99 A0A2A3ENV6 A0A088AAP6 A0A1J1J5Q0 A0A1W4W1Q5 A0A0K8WKW5 E9JBY3 A0A0M9A9W5 A0A1A9X2U9 A0A1I8N4R4 A0A0J7KXG8 A0A2Z2Z004 A0A0L0CEG1 A0A1I8PQ66 K7IWS2 A0A0L7QRF8 B4N6I7 B3M7E5 B4KWS7 B3NF27 Q9VZV2 B4QP18 A0A0A1X9V2 Q29E30 B4PCV6 B4PCV5 A0A3B0J5V4 B4LGX0 U4U9T5 N6U4J9 A0A1B0AD34 B4HTJ6 A0A1A9VSK3 A0A1A9Y2B6 A0A1B0B9H1 A0A0M5IYA4 A0A2P8XG74 A0A069DWM4 A0A182PIM9 A0A182H9P8 A0A212FNA5 B4H407 A0A2S2NVJ0
Pubmed
29372897
26354079
30578597
18362917
19820115
18342250
+ More
24845553 25224926 20966253 12364791 14747013 17210077 20566863 24438588 25244985 20920257 23761445 17510324 25401762 27192063 26823975 20798317 29236932 21719571 21347285 28363844 21282665 25315136 26108605 20075255 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 15632085 17550304 23537049 29403074 26334808 26483478 22118469
24845553 25224926 20966253 12364791 14747013 17210077 20566863 24438588 25244985 20920257 23761445 17510324 25401762 27192063 26823975 20798317 29236932 21719571 21347285 28363844 21282665 25315136 26108605 20075255 17994087 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 15632085 17550304 23537049 29403074 26334808 26483478 22118469
EMBL
KF318218
AGX32025.1
KQ459232
KPJ02577.1
JQ417265
MH899218
+ More
AFI55112.1 AZS52294.1 KX668138 ASM94207.1 MG551526 AUF40393.1 JQ653039 AFM38213.1 NWSH01003324 PCG66555.1 KQ461155 KPJ08355.1 NEVH01016331 PNF25507.1 KQ971361 EFA05323.2 DQ659247 ABG47445.1 GEDC01022730 JAS14568.1 KK852804 KDR16263.1 KM217115 AJO25042.1 AAAB01008807 EAA04006.4 DS235164 EEB12743.1 ATLV01011963 KE524742 KFB36184.1 APCN01003668 AXCM01001639 ADMH02001833 ETN60985.1 AXCN02000560 GGMS01002671 MBY71874.1 GFXV01003929 MBW15734.1 DS232225 EDS37897.1 CH477319 EAT43684.1 GFDL01006125 JAV28920.1 GFDL01006140 JAV28905.1 ABLF02038822 KY576032 AUF41626.1 KY030969 ATU82720.1 KT717329 GBHO01037567 GBRD01008704 GDHC01007234 ANC95003.1 JAG06037.1 JAG57117.1 JAQ11395.1 GL445407 EFN89634.1 KQ981805 KYN35631.1 KY274433 AUM84818.1 KQ982431 KYQ56499.1 GL888424 EGI61144.1 KQ981042 KYN10100.1 AJVK01022918 KQ976716 KYM76839.1 GL439470 EFN67257.1 KQ978344 KYM94854.1 ADTU01002189 ADTU01002190 ADTU01002191 ADTU01002192 ADTU01002193 KY290391 ARE59262.1 KQ434850 KZC08569.1 UFQS01001413 UFQT01001413 SSX10758.1 SSX30440.1 ACPB03000970 GBYB01008986 JAG78753.1 KZ288210 PBC32932.1 CVRI01000067 CRL06798.1 GDHF01031421 GDHF01020037 GDHF01003132 GDHF01000620 JAI20893.1 JAI32277.1 JAI49182.1 JAI51694.1 GL771642 EFZ09665.1 KQ435711 KOX79466.1 LBMM01002318 KMQ94993.1 KY681042 AUP42569.1 JRES01000611 KNC29864.1 AAZX01004127 KQ414784 KOC61139.1 CH964161 EDW79976.2 CH902618 EDV39843.2 CH933809 EDW17524.2 KRG05599.1 KRG05600.1 CH954178 EDV50369.1 AE014296 AY051988 AAF47714.3 AAK93412.1 CM000363 CM002912 EDX09013.1 KMY97240.1 GBXI01006425 GBXI01006211 JAD07867.1 JAD08081.1 CH379070 EAL30233.3 CM000159 EDW93860.2 EDW93859.1 OUUW01000002 SPP77414.1 CH940647 EDW68440.2 KB631935 ERL87331.1 APGK01040030 KB740975 ENN76520.1 CH480817 EDW50267.1 JXJN01010374 CP012525 ALC43111.1 PYGN01002248 PSN31011.1 GBGD01000446 JAC88443.1 JXUM01121179 KQ566471 KXJ70105.1 AGBW02006445 OWR55218.1 CH479208 EDW31122.1 GGMR01008525 MBY21144.1
AFI55112.1 AZS52294.1 KX668138 ASM94207.1 MG551526 AUF40393.1 JQ653039 AFM38213.1 NWSH01003324 PCG66555.1 KQ461155 KPJ08355.1 NEVH01016331 PNF25507.1 KQ971361 EFA05323.2 DQ659247 ABG47445.1 GEDC01022730 JAS14568.1 KK852804 KDR16263.1 KM217115 AJO25042.1 AAAB01008807 EAA04006.4 DS235164 EEB12743.1 ATLV01011963 KE524742 KFB36184.1 APCN01003668 AXCM01001639 ADMH02001833 ETN60985.1 AXCN02000560 GGMS01002671 MBY71874.1 GFXV01003929 MBW15734.1 DS232225 EDS37897.1 CH477319 EAT43684.1 GFDL01006125 JAV28920.1 GFDL01006140 JAV28905.1 ABLF02038822 KY576032 AUF41626.1 KY030969 ATU82720.1 KT717329 GBHO01037567 GBRD01008704 GDHC01007234 ANC95003.1 JAG06037.1 JAG57117.1 JAQ11395.1 GL445407 EFN89634.1 KQ981805 KYN35631.1 KY274433 AUM84818.1 KQ982431 KYQ56499.1 GL888424 EGI61144.1 KQ981042 KYN10100.1 AJVK01022918 KQ976716 KYM76839.1 GL439470 EFN67257.1 KQ978344 KYM94854.1 ADTU01002189 ADTU01002190 ADTU01002191 ADTU01002192 ADTU01002193 KY290391 ARE59262.1 KQ434850 KZC08569.1 UFQS01001413 UFQT01001413 SSX10758.1 SSX30440.1 ACPB03000970 GBYB01008986 JAG78753.1 KZ288210 PBC32932.1 CVRI01000067 CRL06798.1 GDHF01031421 GDHF01020037 GDHF01003132 GDHF01000620 JAI20893.1 JAI32277.1 JAI49182.1 JAI51694.1 GL771642 EFZ09665.1 KQ435711 KOX79466.1 LBMM01002318 KMQ94993.1 KY681042 AUP42569.1 JRES01000611 KNC29864.1 AAZX01004127 KQ414784 KOC61139.1 CH964161 EDW79976.2 CH902618 EDV39843.2 CH933809 EDW17524.2 KRG05599.1 KRG05600.1 CH954178 EDV50369.1 AE014296 AY051988 AAF47714.3 AAK93412.1 CM000363 CM002912 EDX09013.1 KMY97240.1 GBXI01006425 GBXI01006211 JAD07867.1 JAD08081.1 CH379070 EAL30233.3 CM000159 EDW93860.2 EDW93859.1 OUUW01000002 SPP77414.1 CH940647 EDW68440.2 KB631935 ERL87331.1 APGK01040030 KB740975 ENN76520.1 CH480817 EDW50267.1 JXJN01010374 CP012525 ALC43111.1 PYGN01002248 PSN31011.1 GBGD01000446 JAC88443.1 JXUM01121179 KQ566471 KXJ70105.1 AGBW02006445 OWR55218.1 CH479208 EDW31122.1 GGMR01008525 MBY21144.1
Proteomes
UP000053268
UP000218220
UP000053240
UP000235965
UP000007266
UP000027135
+ More
UP000076407 UP000075882 UP000007062 UP000009046 UP000030765 UP000075884 UP000076408 UP000075840 UP000075903 UP000075883 UP000075902 UP000075881 UP000075920 UP000000673 UP000075880 UP000075900 UP000075886 UP000069272 UP000002320 UP000008820 UP000007819 UP000008237 UP000078541 UP000075809 UP000007755 UP000078492 UP000092462 UP000078540 UP000000311 UP000078542 UP000005205 UP000076502 UP000015103 UP000242457 UP000005203 UP000183832 UP000192221 UP000053105 UP000091820 UP000095301 UP000036403 UP000037069 UP000095300 UP000002358 UP000053825 UP000007798 UP000007801 UP000009192 UP000008711 UP000000803 UP000000304 UP000001819 UP000002282 UP000268350 UP000008792 UP000030742 UP000019118 UP000092445 UP000001292 UP000078200 UP000092443 UP000092460 UP000092553 UP000245037 UP000075885 UP000069940 UP000249989 UP000007151 UP000008744
UP000076407 UP000075882 UP000007062 UP000009046 UP000030765 UP000075884 UP000076408 UP000075840 UP000075903 UP000075883 UP000075902 UP000075881 UP000075920 UP000000673 UP000075880 UP000075900 UP000075886 UP000069272 UP000002320 UP000008820 UP000007819 UP000008237 UP000078541 UP000075809 UP000007755 UP000078492 UP000092462 UP000078540 UP000000311 UP000078542 UP000005205 UP000076502 UP000015103 UP000242457 UP000005203 UP000183832 UP000192221 UP000053105 UP000091820 UP000095301 UP000036403 UP000037069 UP000095300 UP000002358 UP000053825 UP000007798 UP000007801 UP000009192 UP000008711 UP000000803 UP000000304 UP000001819 UP000002282 UP000268350 UP000008792 UP000030742 UP000019118 UP000092445 UP000001292 UP000078200 UP000092443 UP000092460 UP000092553 UP000245037 UP000075885 UP000069940 UP000249989 UP000007151 UP000008744
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
U5LUV7
A0A194QAT6
I1VX14
A0A221LFZ8
A0A2H4Z0A2
I6QMM8
+ More
A0A2A4J3B9 A0A194QTM7 A0A2J7QAA7 D2A576 Q0Z942 A0A1B6CMH7 A0A067RC35 A0A0C5CUE1 A0A182WWE3 A0A182L3S0 Q7QIJ1 E0VH87 A0A084VDZ0 A0A182N9E9 A0A182Y814 A0A182I6M1 A0A182VKK7 A0A182LZT7 A0A182TW87 A0A182KCD4 A0A182WBE5 W5JDL8 A0A182IMA2 A0A182RJG7 A0A182QSR0 A0A2S2Q2H3 A0A182FGQ4 A0A2H8TNI6 B0X022 Q17BM9 A0A1Q3FMV9 A0A1Q3FMW1 J9K0Y5 A0A2H4Z2U0 A0A2K8JRQ5 A0A0A9WC72 E2B3W5 A0A195F5G8 A0A2I6Q763 A0A151X836 F4WXH8 A0A195DB08 A0A1B0D2K6 A0A195AY20 E2AH26 A0A195C1V2 A0A158NS63 A0A1V0QBC4 A0A154P9H9 A0A336MKJ0 T1IFA7 A0A0C9RP99 A0A2A3ENV6 A0A088AAP6 A0A1J1J5Q0 A0A1W4W1Q5 A0A0K8WKW5 E9JBY3 A0A0M9A9W5 A0A1A9X2U9 A0A1I8N4R4 A0A0J7KXG8 A0A2Z2Z004 A0A0L0CEG1 A0A1I8PQ66 K7IWS2 A0A0L7QRF8 B4N6I7 B3M7E5 B4KWS7 B3NF27 Q9VZV2 B4QP18 A0A0A1X9V2 Q29E30 B4PCV6 B4PCV5 A0A3B0J5V4 B4LGX0 U4U9T5 N6U4J9 A0A1B0AD34 B4HTJ6 A0A1A9VSK3 A0A1A9Y2B6 A0A1B0B9H1 A0A0M5IYA4 A0A2P8XG74 A0A069DWM4 A0A182PIM9 A0A182H9P8 A0A212FNA5 B4H407 A0A2S2NVJ0
A0A2A4J3B9 A0A194QTM7 A0A2J7QAA7 D2A576 Q0Z942 A0A1B6CMH7 A0A067RC35 A0A0C5CUE1 A0A182WWE3 A0A182L3S0 Q7QIJ1 E0VH87 A0A084VDZ0 A0A182N9E9 A0A182Y814 A0A182I6M1 A0A182VKK7 A0A182LZT7 A0A182TW87 A0A182KCD4 A0A182WBE5 W5JDL8 A0A182IMA2 A0A182RJG7 A0A182QSR0 A0A2S2Q2H3 A0A182FGQ4 A0A2H8TNI6 B0X022 Q17BM9 A0A1Q3FMV9 A0A1Q3FMW1 J9K0Y5 A0A2H4Z2U0 A0A2K8JRQ5 A0A0A9WC72 E2B3W5 A0A195F5G8 A0A2I6Q763 A0A151X836 F4WXH8 A0A195DB08 A0A1B0D2K6 A0A195AY20 E2AH26 A0A195C1V2 A0A158NS63 A0A1V0QBC4 A0A154P9H9 A0A336MKJ0 T1IFA7 A0A0C9RP99 A0A2A3ENV6 A0A088AAP6 A0A1J1J5Q0 A0A1W4W1Q5 A0A0K8WKW5 E9JBY3 A0A0M9A9W5 A0A1A9X2U9 A0A1I8N4R4 A0A0J7KXG8 A0A2Z2Z004 A0A0L0CEG1 A0A1I8PQ66 K7IWS2 A0A0L7QRF8 B4N6I7 B3M7E5 B4KWS7 B3NF27 Q9VZV2 B4QP18 A0A0A1X9V2 Q29E30 B4PCV6 B4PCV5 A0A3B0J5V4 B4LGX0 U4U9T5 N6U4J9 A0A1B0AD34 B4HTJ6 A0A1A9VSK3 A0A1A9Y2B6 A0A1B0B9H1 A0A0M5IYA4 A0A2P8XG74 A0A069DWM4 A0A182PIM9 A0A182H9P8 A0A212FNA5 B4H407 A0A2S2NVJ0
PDB
5WVH
E-value=0,
Score=2413
Ontologies
PATHWAY
GO
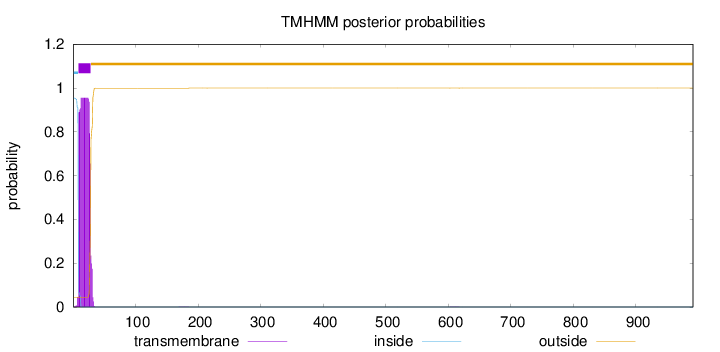
Topology
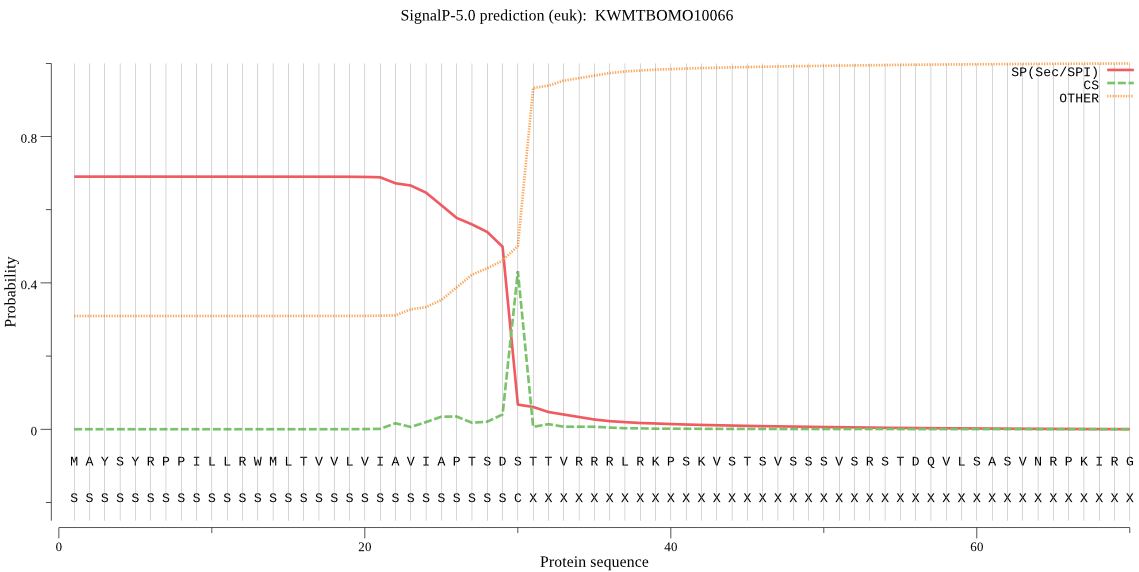
SignalP
Position: 1 - 30,
Likelihood: 0.690327
Length:
991
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.18846
Exp number, first 60 AAs:
19.17803
Total prob of N-in:
0.95508
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 28
outside
29 - 991
Population Genetic Test Statistics
Pi
237.473692
Theta
170.727482
Tajima's D
1.386464
CLR
0.511255
CSRT
0.75781210939453
Interpretation
Uncertain
