Pre Gene Modal
BGIBMGA005537
Annotation
PREDICTED:_inositol_monophosphatase_1_[Amyelois_transitella]
Full name
Inositol-1-monophosphatase
Location in the cell
Cytoplasmic Reliability : 3.881
Sequence
CDS
ATGACTGATAAAGTTGACCAATACTACGACGTTGCTTTGTCCCTGGTAAAAACTTGTGGAAACCTTATTAGTGAGCACATTAATGGTTGTAAATCCTTCGTACTGAAATCATGTGACATAGATCTGGTGACTGAAATAGATAAAAAAGTTGAAGACACACTGGTTGGCGGTCTTTCTAAAGCCTTTCCCGACCACAAGTTTATTGGTGAGGAGTCAGTCGCAGACGGGACGAAATGCGAGTTGACAGACTCACCAACATGGATCATCGATCCAGTTGATGGAACAATGAACTTCGTGCACGGTTTTCCGCACAGCTCAATATCGGTGGGCCTCACAATTAATAAGGAGCCAGTTGTTGGTATTATCTATAATCCTATCTTAGGGCAGATGTTTACTGCTAAGAAAGGTCAAGGTGCCTTCTTAAATGAGACACAAATTCATGTGTCACAAGTTAAAGAGTTTGCCACGGCTTTAGTCACATTTGAAGCAGGTACGAGTCGAGACCCTGAGAGATTTAAGGCTGTGATGGAGAACTATAAGATTCTGGTCAGTAAAGGGCATGGAGTGAGAACATTGGGTTCAGCTGCACTGAATATGGCTATGGTGGCTTTAGGAGGGGCTGATGCTTATTTTGAATTTGGTATTCATGCTTGGGATGTTGCAGCAGGAACTGTTTTGGTGAGGGAAGCTGGTGGAGTGGTCATTGATCCTGCTGGTGGACCATTTGACATATTGTCTAGGAGAATACTCAGTGCCAGTACAATGGAATTAGCAGAAGAGATCTCAAAAAATATAGTTCAGTTTTATCCTGAGAGAGATTAA
Protein
MTDKVDQYYDVALSLVKTCGNLISEHINGCKSFVLKSCDIDLVTEIDKKVEDTLVGGLSKAFPDHKFIGEESVADGTKCELTDSPTWIIDPVDGTMNFVHGFPHSSISVGLTINKEPVVGIIYNPILGQMFTAKKGQGAFLNETQIHVSQVKEFATALVTFEAGTSRDPERFKAVMENYKILVSKGHGVRTLGSAALNMAMVALGGADAYFEFGIHAWDVAAGTVLVREAGGVVIDPAGGPFDILSRRILSASTMELAEEISKNIVQFYPERD
Summary
Catalytic Activity
a myo-inositol phosphate + H2O = myo-inositol + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the inositol monophosphatase superfamily.
Feature
chain Inositol-1-monophosphatase
Uniprot
H9J7P5
A0A2H1WRD3
A0A2A4K9L8
A0A0L7LNG0
A0A212ETH4
A0A0M8ZVL5
+ More
A0A1Y1LV01 A0A026WQU3 E1ZXT3 A0A2A3E2S9 V9IKX9 A0A088A4S7 A0A158NWM2 S4PX24 F4X804 A0A0N1INM7 D7EI01 I4DNT7 N6TA28 A0A151X100 A0A151I1T0 A0A1W4XTE6 K7J8W2 A0A195FXR2 A0A310SEV5 A0A151ID80 A0A2P8Y728 A0A1I8MSK8 A0A151JAJ6 A0A067RKD6 A0A1B6M2Y5 A0A154PLH5 T1PD43 E2BM26 A0A2J7R080 A0A0L7R988 A0A1B6JSF9 A0A182J9P9 A0A232EZ08 U5EW60 B3M6S3 A0A0L0CTI7 A0A182VLE6 A0A182LKH9 Q7PNQ6 A0A182I4F0 A0A182RDH5 A0A1B6MNZ2 Q7KTW5 A0A1B0B572 A0A182WBT6 A0A182X4F0 W5J658 A0A182FFG0 A0A182M3T1 A0A1B6G0D7 A0A1I8P2V6 A0A1A9XWK4 A0A182YJ79 A0A182NIT6 A0A1B6KEZ7 A0A182PDI3 A0A1B0FBN1 B3NIX4 T1H3J9 W8C0S8 D3TPP4 A0A2M4BXJ1 A0A1A9VYM9 A0A2M4AUE6 A0A2M4BWK1 A0A182K2N9 A0A182QYF9 A0A182H9T7 A0A1A9Z3B4 Q17PJ1 A0A1Q3FGP9 A0A1B6K9R1 B4ML52 B4KUM1 E0VZU8 A0A0M3QWI1 A0A1B6KSK4 A0A034W2J6 A0A182TL99 B4ITG8 A0A1W4V9J5 B4QJQ1 A0A0R1E822 B0XAY6 A0A1B6G4A5 A0A0J9RZ74 A0A0K8WKE8 A0A0K8TNH5 A0A3B0KHV9 A0A1B6HCS1 A0A084WMQ2 A0A3B0JT96 E9H383 B5DQB3
A0A1Y1LV01 A0A026WQU3 E1ZXT3 A0A2A3E2S9 V9IKX9 A0A088A4S7 A0A158NWM2 S4PX24 F4X804 A0A0N1INM7 D7EI01 I4DNT7 N6TA28 A0A151X100 A0A151I1T0 A0A1W4XTE6 K7J8W2 A0A195FXR2 A0A310SEV5 A0A151ID80 A0A2P8Y728 A0A1I8MSK8 A0A151JAJ6 A0A067RKD6 A0A1B6M2Y5 A0A154PLH5 T1PD43 E2BM26 A0A2J7R080 A0A0L7R988 A0A1B6JSF9 A0A182J9P9 A0A232EZ08 U5EW60 B3M6S3 A0A0L0CTI7 A0A182VLE6 A0A182LKH9 Q7PNQ6 A0A182I4F0 A0A182RDH5 A0A1B6MNZ2 Q7KTW5 A0A1B0B572 A0A182WBT6 A0A182X4F0 W5J658 A0A182FFG0 A0A182M3T1 A0A1B6G0D7 A0A1I8P2V6 A0A1A9XWK4 A0A182YJ79 A0A182NIT6 A0A1B6KEZ7 A0A182PDI3 A0A1B0FBN1 B3NIX4 T1H3J9 W8C0S8 D3TPP4 A0A2M4BXJ1 A0A1A9VYM9 A0A2M4AUE6 A0A2M4BWK1 A0A182K2N9 A0A182QYF9 A0A182H9T7 A0A1A9Z3B4 Q17PJ1 A0A1Q3FGP9 A0A1B6K9R1 B4ML52 B4KUM1 E0VZU8 A0A0M3QWI1 A0A1B6KSK4 A0A034W2J6 A0A182TL99 B4ITG8 A0A1W4V9J5 B4QJQ1 A0A0R1E822 B0XAY6 A0A1B6G4A5 A0A0J9RZ74 A0A0K8WKE8 A0A0K8TNH5 A0A3B0KHV9 A0A1B6HCS1 A0A084WMQ2 A0A3B0JT96 E9H383 B5DQB3
EC Number
3.1.3.25
Pubmed
19121390
26227816
22118469
28004739
24508170
30249741
+ More
20798317 21347285 23622113 21719571 26354079 18362917 19820115 22651552 23537049 20075255 29403074 25315136 24845553 28648823 17994087 26108605 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25244985 24495485 20353571 26483478 17510324 20566863 25348373 17550304 22936249 26369729 24438588 21292972 15632085
20798317 21347285 23622113 21719571 26354079 18362917 19820115 22651552 23537049 20075255 29403074 25315136 24845553 28648823 17994087 26108605 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 25244985 24495485 20353571 26483478 17510324 20566863 25348373 17550304 22936249 26369729 24438588 21292972 15632085
EMBL
BABH01020381
ODYU01010482
SOQ55623.1
NWSH01000033
PCG80472.1
PCG80473.1
+ More
JTDY01000500 KOB76894.1 AGBW02012580 OWR44793.1 KQ435828 KOX71857.1 GEZM01049775 JAV75855.1 KK107128 QOIP01000006 EZA58400.1 RLU21430.1 GL435089 EFN74004.1 KZ288417 PBC26017.1 JR049446 AEY60969.1 ADTU01028199 GAIX01005783 JAA86777.1 GL888900 EGI57421.1 KQ461156 KPJ08100.1 DS497669 EFA12936.1 AK403133 KQ459232 BAM19577.1 KPJ02569.1 APGK01038392 KB740954 KB632338 ENN77104.1 ERL92908.1 KQ982592 KYQ54074.1 KQ976554 KYM80809.1 AAZX01000812 KQ981193 KYN45097.1 KQ769940 OAD52759.1 KQ977979 KYM98364.1 PYGN01000849 PSN40062.1 KQ979254 KYN22085.1 KK852573 KDR21070.1 GEBQ01009694 JAT30283.1 KQ434938 KZC12080.1 KA646030 AFP60659.1 GL449100 EFN83275.1 NEVH01008279 PNF34251.1 KQ414627 KOC67440.1 GECU01005551 JAT02156.1 NNAY01001586 OXU23517.1 GANO01001576 JAB58295.1 CH902618 EDV39759.2 JRES01000036 KNC34709.1 AAAB01008960 EAA11769.5 APCN01002307 GEBQ01002388 JAT37589.1 AE014296 AY071022 AAF51693.3 AAL48644.1 AAN12162.1 JXJN01008626 ADMH02002078 ETN59471.1 AXCM01002436 GECZ01013908 JAS55861.1 GEBQ01029951 JAT10026.1 CCAG010001559 CH954178 EDV52620.1 CAQQ02373999 GAMC01007299 JAB99256.1 EZ423396 ADD19672.1 GGFJ01008317 MBW57458.1 GGFK01011072 MBW44393.1 GGFJ01008319 MBW57460.1 AXCN02000480 JXUM01069200 JXUM01121422 KQ566499 KQ562540 KXJ70076.1 KXJ75662.1 CH477191 EAT48626.1 GFDL01008326 JAV26719.1 GEBQ01031794 JAT08183.1 CH963847 EDW73110.1 CH933809 EDW18249.1 DS235854 EEB18904.1 CP012525 ALC44189.1 GEBQ01025567 JAT14410.1 GAKP01010386 GAKP01010385 GAKP01010384 JAC48567.1 CH891698 EDW99681.1 CM000363 EDX11351.1 KRK05189.1 DS232603 EDS43956.1 GECZ01012503 GECZ01011595 JAS57266.1 JAS58174.1 CM002912 KMZ00957.1 GDHF01007039 GDHF01000681 JAI45275.1 JAI51633.1 GDAI01001915 JAI15688.1 OUUW01000009 SPP85336.1 GECU01035327 JAS72379.1 ATLV01024500 KE525352 KFB51496.1 SPP85335.1 GL732588 EFX73712.1 CH379069 EDY73857.2
JTDY01000500 KOB76894.1 AGBW02012580 OWR44793.1 KQ435828 KOX71857.1 GEZM01049775 JAV75855.1 KK107128 QOIP01000006 EZA58400.1 RLU21430.1 GL435089 EFN74004.1 KZ288417 PBC26017.1 JR049446 AEY60969.1 ADTU01028199 GAIX01005783 JAA86777.1 GL888900 EGI57421.1 KQ461156 KPJ08100.1 DS497669 EFA12936.1 AK403133 KQ459232 BAM19577.1 KPJ02569.1 APGK01038392 KB740954 KB632338 ENN77104.1 ERL92908.1 KQ982592 KYQ54074.1 KQ976554 KYM80809.1 AAZX01000812 KQ981193 KYN45097.1 KQ769940 OAD52759.1 KQ977979 KYM98364.1 PYGN01000849 PSN40062.1 KQ979254 KYN22085.1 KK852573 KDR21070.1 GEBQ01009694 JAT30283.1 KQ434938 KZC12080.1 KA646030 AFP60659.1 GL449100 EFN83275.1 NEVH01008279 PNF34251.1 KQ414627 KOC67440.1 GECU01005551 JAT02156.1 NNAY01001586 OXU23517.1 GANO01001576 JAB58295.1 CH902618 EDV39759.2 JRES01000036 KNC34709.1 AAAB01008960 EAA11769.5 APCN01002307 GEBQ01002388 JAT37589.1 AE014296 AY071022 AAF51693.3 AAL48644.1 AAN12162.1 JXJN01008626 ADMH02002078 ETN59471.1 AXCM01002436 GECZ01013908 JAS55861.1 GEBQ01029951 JAT10026.1 CCAG010001559 CH954178 EDV52620.1 CAQQ02373999 GAMC01007299 JAB99256.1 EZ423396 ADD19672.1 GGFJ01008317 MBW57458.1 GGFK01011072 MBW44393.1 GGFJ01008319 MBW57460.1 AXCN02000480 JXUM01069200 JXUM01121422 KQ566499 KQ562540 KXJ70076.1 KXJ75662.1 CH477191 EAT48626.1 GFDL01008326 JAV26719.1 GEBQ01031794 JAT08183.1 CH963847 EDW73110.1 CH933809 EDW18249.1 DS235854 EEB18904.1 CP012525 ALC44189.1 GEBQ01025567 JAT14410.1 GAKP01010386 GAKP01010385 GAKP01010384 JAC48567.1 CH891698 EDW99681.1 CM000363 EDX11351.1 KRK05189.1 DS232603 EDS43956.1 GECZ01012503 GECZ01011595 JAS57266.1 JAS58174.1 CM002912 KMZ00957.1 GDHF01007039 GDHF01000681 JAI45275.1 JAI51633.1 GDAI01001915 JAI15688.1 OUUW01000009 SPP85336.1 GECU01035327 JAS72379.1 ATLV01024500 KE525352 KFB51496.1 SPP85335.1 GL732588 EFX73712.1 CH379069 EDY73857.2
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053105
UP000053097
+ More
UP000279307 UP000000311 UP000242457 UP000005203 UP000005205 UP000007755 UP000053240 UP000007266 UP000053268 UP000019118 UP000030742 UP000075809 UP000078540 UP000192223 UP000002358 UP000078541 UP000078542 UP000245037 UP000095301 UP000078492 UP000027135 UP000076502 UP000008237 UP000235965 UP000053825 UP000075880 UP000215335 UP000007801 UP000037069 UP000075903 UP000075882 UP000007062 UP000075840 UP000075900 UP000000803 UP000092460 UP000075920 UP000076407 UP000000673 UP000069272 UP000075883 UP000095300 UP000092443 UP000076408 UP000075884 UP000075885 UP000092444 UP000008711 UP000015102 UP000078200 UP000075881 UP000075886 UP000069940 UP000249989 UP000092445 UP000008820 UP000007798 UP000009192 UP000009046 UP000092553 UP000075902 UP000002282 UP000192221 UP000000304 UP000002320 UP000268350 UP000030765 UP000000305 UP000001819
UP000279307 UP000000311 UP000242457 UP000005203 UP000005205 UP000007755 UP000053240 UP000007266 UP000053268 UP000019118 UP000030742 UP000075809 UP000078540 UP000192223 UP000002358 UP000078541 UP000078542 UP000245037 UP000095301 UP000078492 UP000027135 UP000076502 UP000008237 UP000235965 UP000053825 UP000075880 UP000215335 UP000007801 UP000037069 UP000075903 UP000075882 UP000007062 UP000075840 UP000075900 UP000000803 UP000092460 UP000075920 UP000076407 UP000000673 UP000069272 UP000075883 UP000095300 UP000092443 UP000076408 UP000075884 UP000075885 UP000092444 UP000008711 UP000015102 UP000078200 UP000075881 UP000075886 UP000069940 UP000249989 UP000092445 UP000008820 UP000007798 UP000009192 UP000009046 UP000092553 UP000075902 UP000002282 UP000192221 UP000000304 UP000002320 UP000268350 UP000030765 UP000000305 UP000001819
Pfam
PF00459 Inositol_P
Interpro
CDD
ProteinModelPortal
H9J7P5
A0A2H1WRD3
A0A2A4K9L8
A0A0L7LNG0
A0A212ETH4
A0A0M8ZVL5
+ More
A0A1Y1LV01 A0A026WQU3 E1ZXT3 A0A2A3E2S9 V9IKX9 A0A088A4S7 A0A158NWM2 S4PX24 F4X804 A0A0N1INM7 D7EI01 I4DNT7 N6TA28 A0A151X100 A0A151I1T0 A0A1W4XTE6 K7J8W2 A0A195FXR2 A0A310SEV5 A0A151ID80 A0A2P8Y728 A0A1I8MSK8 A0A151JAJ6 A0A067RKD6 A0A1B6M2Y5 A0A154PLH5 T1PD43 E2BM26 A0A2J7R080 A0A0L7R988 A0A1B6JSF9 A0A182J9P9 A0A232EZ08 U5EW60 B3M6S3 A0A0L0CTI7 A0A182VLE6 A0A182LKH9 Q7PNQ6 A0A182I4F0 A0A182RDH5 A0A1B6MNZ2 Q7KTW5 A0A1B0B572 A0A182WBT6 A0A182X4F0 W5J658 A0A182FFG0 A0A182M3T1 A0A1B6G0D7 A0A1I8P2V6 A0A1A9XWK4 A0A182YJ79 A0A182NIT6 A0A1B6KEZ7 A0A182PDI3 A0A1B0FBN1 B3NIX4 T1H3J9 W8C0S8 D3TPP4 A0A2M4BXJ1 A0A1A9VYM9 A0A2M4AUE6 A0A2M4BWK1 A0A182K2N9 A0A182QYF9 A0A182H9T7 A0A1A9Z3B4 Q17PJ1 A0A1Q3FGP9 A0A1B6K9R1 B4ML52 B4KUM1 E0VZU8 A0A0M3QWI1 A0A1B6KSK4 A0A034W2J6 A0A182TL99 B4ITG8 A0A1W4V9J5 B4QJQ1 A0A0R1E822 B0XAY6 A0A1B6G4A5 A0A0J9RZ74 A0A0K8WKE8 A0A0K8TNH5 A0A3B0KHV9 A0A1B6HCS1 A0A084WMQ2 A0A3B0JT96 E9H383 B5DQB3
A0A1Y1LV01 A0A026WQU3 E1ZXT3 A0A2A3E2S9 V9IKX9 A0A088A4S7 A0A158NWM2 S4PX24 F4X804 A0A0N1INM7 D7EI01 I4DNT7 N6TA28 A0A151X100 A0A151I1T0 A0A1W4XTE6 K7J8W2 A0A195FXR2 A0A310SEV5 A0A151ID80 A0A2P8Y728 A0A1I8MSK8 A0A151JAJ6 A0A067RKD6 A0A1B6M2Y5 A0A154PLH5 T1PD43 E2BM26 A0A2J7R080 A0A0L7R988 A0A1B6JSF9 A0A182J9P9 A0A232EZ08 U5EW60 B3M6S3 A0A0L0CTI7 A0A182VLE6 A0A182LKH9 Q7PNQ6 A0A182I4F0 A0A182RDH5 A0A1B6MNZ2 Q7KTW5 A0A1B0B572 A0A182WBT6 A0A182X4F0 W5J658 A0A182FFG0 A0A182M3T1 A0A1B6G0D7 A0A1I8P2V6 A0A1A9XWK4 A0A182YJ79 A0A182NIT6 A0A1B6KEZ7 A0A182PDI3 A0A1B0FBN1 B3NIX4 T1H3J9 W8C0S8 D3TPP4 A0A2M4BXJ1 A0A1A9VYM9 A0A2M4AUE6 A0A2M4BWK1 A0A182K2N9 A0A182QYF9 A0A182H9T7 A0A1A9Z3B4 Q17PJ1 A0A1Q3FGP9 A0A1B6K9R1 B4ML52 B4KUM1 E0VZU8 A0A0M3QWI1 A0A1B6KSK4 A0A034W2J6 A0A182TL99 B4ITG8 A0A1W4V9J5 B4QJQ1 A0A0R1E822 B0XAY6 A0A1B6G4A5 A0A0J9RZ74 A0A0K8WKE8 A0A0K8TNH5 A0A3B0KHV9 A0A1B6HCS1 A0A084WMQ2 A0A3B0JT96 E9H383 B5DQB3
PDB
6GJ0
E-value=8.78591e-72,
Score=685
Ontologies
PATHWAY
GO
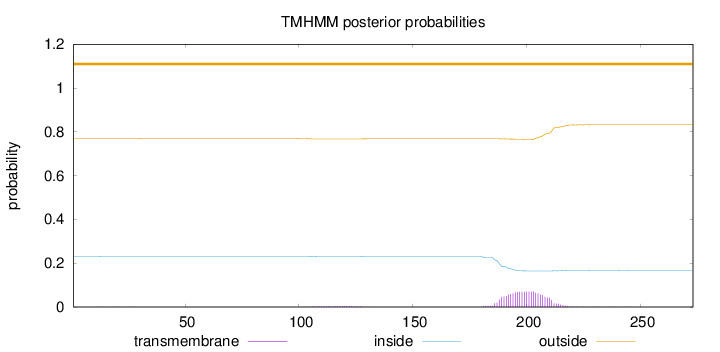
Topology
Length:
273
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.62798
Exp number, first 60 AAs:
0.004
Total prob of N-in:
0.23111
outside
1 - 273
Population Genetic Test Statistics
Pi
166.470227
Theta
178.407159
Tajima's D
-0.203281
CLR
34.835037
CSRT
0.314084295785211
Interpretation
Uncertain
