Gene
KWMTBOMO10056 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005536
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_myosin_heavy_chain?_non-muscle-like_[Plutella_xylostella]
Full name
Myosin heavy chain, non-muscle
Alternative Name
Myosin II
Non-muscle MHC
Zipper protein
Non-muscle MHC
Zipper protein
Location in the cell
Cytoskeletal Reliability : 3.499
Sequence
CDS
ATGGTGGACATGGATAGAAACGACCCGGAGCTCCGGTATCTGTCGGTGGATAGAAACAGCTTCAATGATCCAGCTACGCAGGCGGAATGGACGCAGAAGAGGTTAGTTTGGGTGCCTCACGAGAACCAAGGATTCGTAGCGGCCGGAATCAAAGGAGAGCGAGGAGATGAAGTTGAAGTGGAGATAGCCGAGTCAGGAAAGCGGATGTTGGTGCCCATAGATGATATCCAGAAAATGAATCCACCAAAGTTCGATAAAGTGGAGGATATGGCTGAACTGACCTGCTTAAACGAGGCTTCTGTTCTGCACAACATAAAAGACAGATACTATTCTGGACTCATTTATACCTACTCTGGCCTCTTCTGTGTGGTCGTCAATCCGTACAAGAAGTTGCCGATATACACGGAGAAGATCATGGAGAGATACAAAGGAATCAAGAGGCATGAAGTACCTCCACACGTTTTCGCCATCACCGACACGGCGTATCGCTCCATGCTTCAAGATCGCGAAGACCAATCCATACTCTGCACCGGAGAGTCGGGCGCTGGTAAAACGGAGAACACCAAAAAGGTCATACAGTACTTAGCGTACGTCGCCGCTTCGAAACCTAAAGGATCAGGAGCGGGACCGTCGCCACAGCTGATAATAGGCGAGTTGGAACAGCAATTGTTGCAGGCCAATCCGATATTGGAGGCGTTCGGTAATGCGAAGACGGTCAAAAACGATAACTCATCTAGATTCGGTAAATTCATAAGGATAAACTTCGACGCGTCCGGCTTCATAGCCGGTGCCAACATCGAAACGTATCTATTAGAAAAATCGAGGGCCATAAGACAAGCGAAGGACGAGAGGACATTCCACATTTTCTATCAGCTGCTGACCGGAGCGACCCCGCAGCAGAAGGCCGAGTTCATTCTAGAAGATGCGAAAAGCTACCCGTTTTTATCTAGTGGCGCTCTACCGGTGCCCGGCATCGATGACGCGGCCGAGTACCAGGCCACCATGAAGTCAATGAACATCATGGGCATGAACAACGAGGACTTCAATTCGATATTTAGGATAGTCTCGGCGGTTCTCTTGTTCGGGTCGATGCAGTTCAAGCAGGAAAGGAATTCGGATCAAGCCACGTTGCCCGACAACACCGTGGCGCAGAAAATCGCGCATTTACTTGGTCTTTCCGTAACGGAAATGACGAAGGCTTTCTTGAAACCGAGAATTAAAGTGGGTCGCGATTTCGTCACAAAAGCACAAACCAAGGAACAAGTTGAGTTTTCTGTTGAAGCTATCGGTAAGGCTTGTTACGAACGTTTGTTCAGATGGCTGGTGAACAGGATCAACAGGTCTCTGGACAGGACGAAGAGACAAGGCGCCTCGTTTATCGGCATCCTCGACATGGCAGGGTTCGAAATCTTCGAATTGAACTCCTTCGAACAGCTCTGCATCAACTACACGAACGAGAAGTTGCAGCAACTCTTCAACCACACTATGTTTATATTGGAACAGGAAGAGTATCAGCGCGAAGGAATCGAATGGAAATTCATTGATTTCGGTCTCGATCTCCAACCGACCATCGATCTCATCGACAAACCCATGGGCATCATGGCCTTGCTCGATGAAGAATGTTGGTTCCCCAAGGCGACAGACAAGACCTTCGTCGAGAAGTTGGTGTCGGCCCACTCCGTGCACCCGAAGTTTATGAAGACTGACTTCCGCGGCGTGGCAGACTTCGCCATCATTCACTACGCCGGTAAAGTGGACTACTCAGCCGCCCAGTGGCTCATGAAGAACATGGACCCGCTCAACGAGAACGTCGTGTCGCTTCTTCAGAACTCGCAGGATCCTTTCGTTTGTCACATCTGGAAGGACGCTGAGATTGTCGGCATGGCGCAGCAGGCGATGACCGACACGCAGTTCGGTGCAAGAACGCGCAAAGGAATGTTCAGGACTGTCTCCCAACTGTATAAGGAGCAGTTGACTAAACTGATGGCCACATTGAGGAACACCAATCCGAACTTTGTGAGGTGCATCATTCCCAACCATGAAAAGAAAGCAGGAAAGATTGAGGCGCCCCTCGTCTTGGACCAGTTGCGGTGCAACGGTGTCCTCGAGGGCATCAGAATTTGCAGACAGGGCTTCCCGAACAGGATTCCGTTCCAAGAGTTCAGGCAGCGCTATGAACTCCTCACTCCGAACATCATTCCTAAAGGTTTCATGGACGGAAAGAAGGCCTGCGAACAGATGATCGAGGCTCTCGAGCTGGACCACAATCTGTACAGAGTTGGTCAGTCGAAAATATTCTTCAGGGCCGGTGTGCTGGCTCACCTGGAAGAGGAGAGGGATTACAAGATCACGGACTTGATAGTCAACTTCCAGGCGTTCTGCCGCGGCTACCTGGCCCGGCGGAACTACCAGAAGAGACTCCAGCAACTCAACGCCATTCGGATCATTCAGAGGAACTGCGCTGCGTATCTCAAGTTGAGGAATTGGCAGTGGTGGAGACTGTATATTAAGGTGAAACCCCTTCTAGAAGTAACGAAACAAGAAGAGAAACTCACCCAGAAAGAGGATGAGCTACGTCAGATACGCGAGAGACTCGAAACCCAGGCGAAACGGTCCCAGGAGTTGGAGCAGAAGTACCAGCAGGCTACCGCTGAGAAGACCCAGCTCGCCGAACAATTGCAGGCGGAATTGGAACTGTGCGCCGAAGCGGAAGAGAGTCGAGCTCGCGCCGCGGCCAGGAAGCAGGAGCTGGAGGAGCTGCTGCACGACCTCGAGGGACGGATAGAGGAGGAGGAGGAGAGATGCAATGCTCTGCAGCAGGAGAAGAAACGTCTCCAGCTGAATATACAGGACCTGGAGGAACAGCTGGAAGAAGAAGAGGCGGCCCGTCAAAAGTTACAGCTGGAGCGTGTCCAGTGTGACTCCAAGATCAAGAAGCTGGAGGAGGACCTCGCCCTGAGCGATGACACCAACCAGAAACTCGTCAAGGAGAAGAAGATTTTGGAAGAACGGGCCAACGATTTGTCTCAAGCGTTGGCCGAAGAAGAAGAGAAGGCCAAACATTTGAGCAAACTCAAGGCCAAACAGGAGTCTGCTATCGCCGAACTTGAAGAGAAATTACTTAAGGATCACCAAATGCGACAAGAAGTGGACCGAGCCAAGAGGAAACTGGAGACGGAGCTGCAGGACGTGAAGGAGCAGCTCGTCGAGAGGAAGGCGCAGCTCGAGGAGCTGCAGATCGCACTCGTGAAGCGCGAGGAGGAGGTGACGGCGGCGGCGGCGCGCGCGGAGGCGGAGTCGGCCGCGCGCGCCGCCGCGCAGAAGGCCGCGCGCGAGGCGGAGTCGGCGCTGGCCGACCTGCACGACGACCTGGACGCCGAGCGCGCCGCCCGCTCCAAGGCAGAGAAGCTGCGCAGGGACCTCAACGAGGAGCTCGAGGCGCTCAAGAGCGAGCTGCTCGACTCGCTGGACACCACCGCAGCCCAACAGGAGCTCCGGTCGAAGCGCGAGCAGGAGGTGGCGGTGCTGAAGCGCAGCCTGGAGGAGGAGGCGGCGGCGCACGAGGCGGCGCTCACGCAGCAGCGCCACAAGCACTCGCTCGACCTGCAGGCGCTCAACGAGCAGCAGGAACAACTCAAAAAGGCTAAAGCTGCTCTTGAGAAAGCGAAGCAGTCGCTGGAAGCAGAAAACGCGGACCTCGCCACGGAACTGAAGTCCGCCAGCGCCGCCCGCGCCGACGGCGAGAAGAAACGGAAGCAGGCGGAGGCCCAGCTCGCCGACCTCGCCGCCAAGCTGCAGGACATCGAGAGGACCAGAGCGGAAGTCCAAGAGAAATGCGTCCGTCTGCAGAGCGAGGCGGAGCAGGCCGCGGCTCAGCTGGAGGACGCCGAGCTGAAAGCCTCCGCCGCCGTCAAGCAGTCCGCTACCATCGGCGCGCAGCTCACGGAGGCGCAGGCACTTTTGGAAGAAGAAACGAAGCAGAAGCTGTCCCTCCAGACGAAGCTCCGCAACATTGAACAAGAGTTGGAACAGGCGCGCGATCAGCTCGAAGAAGAAGAGGAGGCTAAGAAAAATCTCGAGAAACAGGTGACGGCGCTAACGGTCCAAGTGTCCGAGGCGAAGAAGAAAGCAGAGGAGGAAGCTAAGAATGCGGCCGCCCTGGAGGAACAGCGCAAGAAGCTCAGCAAGGACGTGGAAGCTCTGCACCGGCAGATCGACGAGCTGCAGCAGGCCAACGACAAACTAGACAAGAGTAAAAAGAAACTTCAAGCCGAGCTGGAAGACACTAACATAGAACTAGAAGCCCAGCGAGCCAAAGTTATGGAGCTTGAGAAGAAACAGAAGAGCTTCGATAAGGTGGTGGCCGAAGAGCGTGCCGTGGCGGAGCGGTACGCCGCCGAGAGAGATCAGGCGGAGCACGAGGCCAGGGAGAAGGAGACCAGGGTGCTCTCTCTGACCAGGGAATTAGATGATGCTGCCGAGAAGATCGAAGAGCTAGAACGGACGAAGCGAGTGCTCCAAGCGGAGCTGGACGAGCTGGCCAACTCGCAAGGCACGGCCGACAAGAACGTCCACGAGCTGGAGCGAGCCAAGCGAGCGCTGGAGTCGCAGCTGGCCGAGCTGCACGCGCAGAACGAGGAGATCGAGGACGACCTGCAGCTCACCGAGGACGCCAAGCTGCGGCTCGAGGTCAACATGCAGGCCATGCGCGCGCAGTTCGAGCGCGACCTGCAGGCTAAAGAAGAACAAGGCGAGGAGAAGCGTCGCGGCATCGTGAAGCAGCTGCGCGACGTGGAGACGGAGCTGGAGGAGGAGCGCAAGCAGCGAGCCGCCGCGCTCGCGCAGAGGAAGAAGCTCGAGGCCGACCTCAAGGACGCCGAGCAGGCGCTGCACCTCGCCAACAAGGTCAAAGAAGATGCCGCAAAACAGGTTAAGAAACTACAGCAGCAGCTCAAGGAAGCTGCCCGCGAGGCCGAAGAAGCCAGAGCCGGTCGCGAAGAGGCTTCGGCCTCGGCCAGAGAAGCCGAGAGGAAGCTGAAGGCCCTGGAAGCCGAGGCTCTCCATCACGTCGAAGAGCTGGCGGCCGCCGAGCGAGCGAGACGACAGGCTGAAGCAGAGAGAGACGACCTGCACGATGAACTCAATAATACCTCCGCTAAGAGTACACTTCTGATCGATGAGAAGAAACGTCTCGAGGCCAGAATAGCCGCCCTCGAGGAGGACATCGACGAAGAACAATCCAACAACGAAATACTTAGCGATAGACTTAGGAAGTCACAGAACCAAATCGATACATTAACAATGGAGTTGGGAACAGAGAAATCTGCAACACAGAAGCTGGAGAGCGGCAAACTCGTCCTCGAGAGGCAGAACAAGGAGCTGAAGGCGAAACTCGCTGAAATGGAGACGGCGGGACGGACCAAGACCAAGGGTGTTATTACGTCATTGGAGCTGAAGGTGGCCAACCTCGAGGAGCAACTGGAGGCGGAGTCGAGAGAGCGCCTCGCTCAACAGAAGGCCTCGAGGAAACTCGATAAGAAGATGAAGGAGTTGGCTCTTCAACTCGACGAGGAAAGAAGACAGGCCGACCAGTTCAAGGAACAGATTGAAAAGATGAACGTGAGAATGAAAACACTAAAGCGTCGTTTAGACGAGGCGGAAGAGGAAGTGCAGCGCGAGAGGGCCGGCAAGAGGAAGGCGCAGAGGGAACTCGAGGACCTCAACGAGGCTCACGAAACCGTCACCCTCGAGTGCAACAACTTGAGGAACAAACTCAGGCGACAAGGTGGGCCGATAAGTCTGTCCAGCGGCTCCACGACCTCGCGCAGGAACCGTCCATCCTTGGCTCCGGGAGGCGGCTCCGGAGACGAGTCCTTCGATGACGTCAACGATACAACAAACAGCGTCGAGTAA
Protein
MVDMDRNDPELRYLSVDRNSFNDPATQAEWTQKRLVWVPHENQGFVAAGIKGERGDEVEVEIAESGKRMLVPIDDIQKMNPPKFDKVEDMAELTCLNEASVLHNIKDRYYSGLIYTYSGLFCVVVNPYKKLPIYTEKIMERYKGIKRHEVPPHVFAITDTAYRSMLQDREDQSILCTGESGAGKTENTKKVIQYLAYVAASKPKGSGAGPSPQLIIGELEQQLLQANPILEAFGNAKTVKNDNSSRFGKFIRINFDASGFIAGANIETYLLEKSRAIRQAKDERTFHIFYQLLTGATPQQKAEFILEDAKSYPFLSSGALPVPGIDDAAEYQATMKSMNIMGMNNEDFNSIFRIVSAVLLFGSMQFKQERNSDQATLPDNTVAQKIAHLLGLSVTEMTKAFLKPRIKVGRDFVTKAQTKEQVEFSVEAIGKACYERLFRWLVNRINRSLDRTKRQGASFIGILDMAGFEIFELNSFEQLCINYTNEKLQQLFNHTMFILEQEEYQREGIEWKFIDFGLDLQPTIDLIDKPMGIMALLDEECWFPKATDKTFVEKLVSAHSVHPKFMKTDFRGVADFAIIHYAGKVDYSAAQWLMKNMDPLNENVVSLLQNSQDPFVCHIWKDAEIVGMAQQAMTDTQFGARTRKGMFRTVSQLYKEQLTKLMATLRNTNPNFVRCIIPNHEKKAGKIEAPLVLDQLRCNGVLEGIRICRQGFPNRIPFQEFRQRYELLTPNIIPKGFMDGKKACEQMIEALELDHNLYRVGQSKIFFRAGVLAHLEEERDYKITDLIVNFQAFCRGYLARRNYQKRLQQLNAIRIIQRNCAAYLKLRNWQWWRLYIKVKPLLEVTKQEEKLTQKEDELRQIRERLETQAKRSQELEQKYQQATAEKTQLAEQLQAELELCAEAEESRARAAARKQELEELLHDLEGRIEEEEERCNALQQEKKRLQLNIQDLEEQLEEEEAARQKLQLERVQCDSKIKKLEEDLALSDDTNQKLVKEKKILEERANDLSQALAEEEEKAKHLSKLKAKQESAIAELEEKLLKDHQMRQEVDRAKRKLETELQDVKEQLVERKAQLEELQIALVKREEEVTAAAARAEAESAARAAAQKAAREAESALADLHDDLDAERAARSKAEKLRRDLNEELEALKSELLDSLDTTAAQQELRSKREQEVAVLKRSLEEEAAAHEAALTQQRHKHSLDLQALNEQQEQLKKAKAALEKAKQSLEAENADLATELKSASAARADGEKKRKQAEAQLADLAAKLQDIERTRAEVQEKCVRLQSEAEQAAAQLEDAELKASAAVKQSATIGAQLTEAQALLEEETKQKLSLQTKLRNIEQELEQARDQLEEEEEAKKNLEKQVTALTVQVSEAKKKAEEEAKNAAALEEQRKKLSKDVEALHRQIDELQQANDKLDKSKKKLQAELEDTNIELEAQRAKVMELEKKQKSFDKVVAEERAVAERYAAERDQAEHEAREKETRVLSLTRELDDAAEKIEELERTKRVLQAELDELANSQGTADKNVHELERAKRALESQLAELHAQNEEIEDDLQLTEDAKLRLEVNMQAMRAQFERDLQAKEEQGEEKRRGIVKQLRDVETELEEERKQRAAALAQRKKLEADLKDAEQALHLANKVKEDAAKQVKKLQQQLKEAAREAEEARAGREEASASAREAERKLKALEAEALHHVEELAAAERARRQAEAERDDLHDELNNTSAKSTLLIDEKKRLEARIAALEEDIDEEQSNNEILSDRLRKSQNQIDTLTMELGTEKSATQKLESGKLVLERQNKELKAKLAEMETAGRTKTKGVITSLELKVANLEEQLEAESRERLAQQKASRKLDKKMKELALQLDEERRQADQFKEQIEKMNVRMKTLKRRLDEAEEEVQRERAGKRKAQRELEDLNEAHETVTLECNNLRNKLRRQGGPISLSSGSTTSRRNRPSLAPGGGSGDESFDDVNDTTNSVE
Summary
Description
Nonmuscle myosin appears to be responsible for cellularization. Required for morphogenesis and cytokinesis (PubMed:24786584). Necessary for auditory transduction: plays a role in Johnston's organ organization by acting in scolopidial apical attachment (PubMed:27331610). Interaction with the myosin ck may be important for this function (PubMed:27331610).
Subunit
Interacts with sau (PubMed:24786584). Interacts with ck and Ubr3 (PubMed:27331610).
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Keywords
Actin-binding
Alternative splicing
ATP-binding
Calmodulin-binding
Cell projection
Coiled coil
Complete proteome
Motor protein
Myosin
Nucleotide-binding
Phosphoprotein
Reference proteome
Ubl conjugation
Feature
chain Myosin heavy chain, non-muscle
splice variant In isoform 2 and isoform 4.
splice variant In isoform 2 and isoform 4.
Uniprot
A0A1E1WTX5
B4LL10
A0A0Q9WGS5
A0A1W4XRS5
A0A1W4XQX5
A0A1W4XGR0
+ More
A0A2H8TR98 J9JUV1 A0A348G639 A0A0J7L5I2 A0A151X7M5 A0A195CBE9 A0A2A3E894 A0A088AJL9 A0A0C9R8B0 A0A158NA72 A0A195FR87 A0A195DMW3 A0A195BKD8 A0A3L8D615 A0A0L7QUX8 A0A1Q3FU39 A0A1I8JT87 A0A1B6BZK7 A0A2M4B8B1 A0A2M3ZY98 A0A2M3YXA0 A0A2M3YXA1 A0A1B6CXQ7 A0A182VCA9 A0A194QAS5 A0A0N0U341 A0A182QX48 A0A3S2NF93 W8AZY5 A0A0A1XSI3 A0A0K8WES0 A0A0K8TZ85 A0A1S4EYP3 A0A034W2L7 W4VRK7 V5JDF9 A0A0Q9X852 A0A1I8MRB0 T1PEM7 A0A0P9BZA8 A0A1I8NSL1 A0A0Q9X8Z6 A0A1L8E5H3 A0A0Q9XKV7 A0A1L8E5N4 A0A232EVU4 A0A0Q9WPB9 A0A0Q9WXH9 B4MJY8 A0A0M4EI37 A0A1W4V0I6 E0VNT1 A0A0Q5W3C6 A0A0J9RM65 Q59E58 A0A0R1DTW1 Q59E59 A0A1W4ULZ8 A0A0B4K7Q4 A0A0B4JD95 A0A0J9RL63 A0A0B4JD57 A0A224XCL1 A0A0J9RKL7 A0A0V0G429 A0A069DXW8 A0A146LQ23 A0A0R3NL29 A0A146MEA6 A0A146LT94 A0A1W4XRY8 A0A3B0J4P3 A0A2J7QWP2 A0A1W7RA51 Q99323-2 Q99323-1 W5JE10 A0A139WCQ8 A0A1Y1LEQ6 A0A1Y1LAZ4 V5GNY8 A0A0P5J9N0 A0A164ZVZ4 A0A0P5VW45 A0A0P5KU35 A0A0P5IY09 A0A0N8DH95 A0A0P6F3T2 A0A0P5UQL1 A0A0P5X1M2 A0A0P5SAS7 A0A0P5PUF7 A0A1W7RA82
A0A2H8TR98 J9JUV1 A0A348G639 A0A0J7L5I2 A0A151X7M5 A0A195CBE9 A0A2A3E894 A0A088AJL9 A0A0C9R8B0 A0A158NA72 A0A195FR87 A0A195DMW3 A0A195BKD8 A0A3L8D615 A0A0L7QUX8 A0A1Q3FU39 A0A1I8JT87 A0A1B6BZK7 A0A2M4B8B1 A0A2M3ZY98 A0A2M3YXA0 A0A2M3YXA1 A0A1B6CXQ7 A0A182VCA9 A0A194QAS5 A0A0N0U341 A0A182QX48 A0A3S2NF93 W8AZY5 A0A0A1XSI3 A0A0K8WES0 A0A0K8TZ85 A0A1S4EYP3 A0A034W2L7 W4VRK7 V5JDF9 A0A0Q9X852 A0A1I8MRB0 T1PEM7 A0A0P9BZA8 A0A1I8NSL1 A0A0Q9X8Z6 A0A1L8E5H3 A0A0Q9XKV7 A0A1L8E5N4 A0A232EVU4 A0A0Q9WPB9 A0A0Q9WXH9 B4MJY8 A0A0M4EI37 A0A1W4V0I6 E0VNT1 A0A0Q5W3C6 A0A0J9RM65 Q59E58 A0A0R1DTW1 Q59E59 A0A1W4ULZ8 A0A0B4K7Q4 A0A0B4JD95 A0A0J9RL63 A0A0B4JD57 A0A224XCL1 A0A0J9RKL7 A0A0V0G429 A0A069DXW8 A0A146LQ23 A0A0R3NL29 A0A146MEA6 A0A146LT94 A0A1W4XRY8 A0A3B0J4P3 A0A2J7QWP2 A0A1W7RA51 Q99323-2 Q99323-1 W5JE10 A0A139WCQ8 A0A1Y1LEQ6 A0A1Y1LAZ4 V5GNY8 A0A0P5J9N0 A0A164ZVZ4 A0A0P5VW45 A0A0P5KU35 A0A0P5IY09 A0A0N8DH95 A0A0P6F3T2 A0A0P5UQL1 A0A0P5X1M2 A0A0P5SAS7 A0A0P5PUF7 A0A1W7RA82
Pubmed
17994087
21347285
30249741
26354079
24495485
25830018
+ More
25348373 25315136 28648823 20566863 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26334808 26823975 15632085 2117279 8568878 18327897 24786584 27331610 20920257 23761445 18362917 19820115 28004739
25348373 25315136 28648823 20566863 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26334808 26823975 15632085 2117279 8568878 18327897 24786584 27331610 20920257 23761445 18362917 19820115 28004739
EMBL
GDQN01000758
JAT90296.1
CH940648
EDW61817.2
KRF80254.1
GFXV01004919
+ More
MBW16724.1 ABLF02028715 FX985578 BBF97912.1 LBMM01000492 KMQ98222.1 KQ982446 KYQ56324.1 KQ978009 KYM98127.1 KZ288349 PBC27426.1 GBYB01004910 GBYB01008414 GBYB01009122 JAG74677.1 JAG78181.1 JAG78889.1 ADTU01010054 ADTU01010055 ADTU01010056 KQ981305 KYN42978.1 KQ980734 KYN13839.1 KQ976455 KYM85152.1 QOIP01000012 RLU15912.1 KQ414730 KOC62443.1 GFDL01003931 JAV31114.1 APCN01000207 GEDC01030849 JAS06449.1 GGFJ01000148 MBW49289.1 GGFK01000150 MBW33471.1 GGFM01000129 MBW20880.1 GGFM01000134 MBW20885.1 GEDC01019265 JAS18033.1 KQ459232 KPJ02567.1 KQ435919 KOX68708.1 AXCN02000377 RSAL01000062 RVE49552.1 GAMC01016127 JAB90428.1 GBXI01014719 GBXI01013283 GBXI01002443 GBXI01000779 JAC99572.1 JAD01009.1 JAD11849.1 JAD13513.1 GDHF01017819 GDHF01002658 JAI34495.1 JAI49656.1 GDHF01032721 GDHF01003672 JAI19593.1 JAI48642.1 GAKP01010602 GAKP01010601 JAC48350.1 GANO01001870 JAB58001.1 KC122900 AGI96978.1 CH933808 KRG04566.1 KA646590 AFP61219.1 CH902619 KPU76661.1 KRG04565.1 GFDF01000131 JAV13953.1 KRG04568.1 GFDF01000130 JAV13954.1 NNAY01001953 OXU22470.1 CH963846 KRF97567.1 KRF97566.1 EDW72427.2 CP012524 ALC42372.1 DS235346 EEB15037.1 CH954179 KQS63167.1 CM002911 KMY96509.1 AE013599 AAX52688.1 ADV37253.1 ADV37255.1 CM000158 KRK00619.1 AAX52687.1 AFH08269.1 ADV37256.1 KMY96512.1 ADV37254.1 GFTR01008918 JAW07508.1 KMY96508.1 GECL01003329 JAP02795.1 GBGD01000041 JAC88848.1 GDHC01009743 JAQ08886.1 CM000071 KRT01625.1 GDHC01001297 JAQ17332.1 GDHC01007658 JAQ10971.1 OUUW01000001 SPP74472.1 NEVH01009422 PNF33006.1 GFAH01000364 JAV48025.1 M35012 U35816 ADMH02001422 ETN62597.1 KQ971362 KYB25729.1 GEZM01060901 JAV70830.1 GEZM01060902 JAV70829.1 GALX01005149 JAB63317.1 GDIQ01205035 GDIQ01205033 GDIQ01205031 JAK46694.1 LRGB01000687 KZS16848.1 GDIP01094325 JAM09390.1 GDIQ01205036 GDIQ01205034 GDIQ01205032 JAK46689.1 GDIQ01207828 GDIQ01024405 JAK43897.1 GDIP01033887 JAM69828.1 GDIQ01053392 JAN41345.1 GDIP01127067 JAL76647.1 GDIP01090770 JAM12945.1 GDIQ01196102 GDIQ01096169 JAL55557.1 GDIQ01123210 JAL28516.1 GFAH01000363 JAV48026.1
MBW16724.1 ABLF02028715 FX985578 BBF97912.1 LBMM01000492 KMQ98222.1 KQ982446 KYQ56324.1 KQ978009 KYM98127.1 KZ288349 PBC27426.1 GBYB01004910 GBYB01008414 GBYB01009122 JAG74677.1 JAG78181.1 JAG78889.1 ADTU01010054 ADTU01010055 ADTU01010056 KQ981305 KYN42978.1 KQ980734 KYN13839.1 KQ976455 KYM85152.1 QOIP01000012 RLU15912.1 KQ414730 KOC62443.1 GFDL01003931 JAV31114.1 APCN01000207 GEDC01030849 JAS06449.1 GGFJ01000148 MBW49289.1 GGFK01000150 MBW33471.1 GGFM01000129 MBW20880.1 GGFM01000134 MBW20885.1 GEDC01019265 JAS18033.1 KQ459232 KPJ02567.1 KQ435919 KOX68708.1 AXCN02000377 RSAL01000062 RVE49552.1 GAMC01016127 JAB90428.1 GBXI01014719 GBXI01013283 GBXI01002443 GBXI01000779 JAC99572.1 JAD01009.1 JAD11849.1 JAD13513.1 GDHF01017819 GDHF01002658 JAI34495.1 JAI49656.1 GDHF01032721 GDHF01003672 JAI19593.1 JAI48642.1 GAKP01010602 GAKP01010601 JAC48350.1 GANO01001870 JAB58001.1 KC122900 AGI96978.1 CH933808 KRG04566.1 KA646590 AFP61219.1 CH902619 KPU76661.1 KRG04565.1 GFDF01000131 JAV13953.1 KRG04568.1 GFDF01000130 JAV13954.1 NNAY01001953 OXU22470.1 CH963846 KRF97567.1 KRF97566.1 EDW72427.2 CP012524 ALC42372.1 DS235346 EEB15037.1 CH954179 KQS63167.1 CM002911 KMY96509.1 AE013599 AAX52688.1 ADV37253.1 ADV37255.1 CM000158 KRK00619.1 AAX52687.1 AFH08269.1 ADV37256.1 KMY96512.1 ADV37254.1 GFTR01008918 JAW07508.1 KMY96508.1 GECL01003329 JAP02795.1 GBGD01000041 JAC88848.1 GDHC01009743 JAQ08886.1 CM000071 KRT01625.1 GDHC01001297 JAQ17332.1 GDHC01007658 JAQ10971.1 OUUW01000001 SPP74472.1 NEVH01009422 PNF33006.1 GFAH01000364 JAV48025.1 M35012 U35816 ADMH02001422 ETN62597.1 KQ971362 KYB25729.1 GEZM01060901 JAV70830.1 GEZM01060902 JAV70829.1 GALX01005149 JAB63317.1 GDIQ01205035 GDIQ01205033 GDIQ01205031 JAK46694.1 LRGB01000687 KZS16848.1 GDIP01094325 JAM09390.1 GDIQ01205036 GDIQ01205034 GDIQ01205032 JAK46689.1 GDIQ01207828 GDIQ01024405 JAK43897.1 GDIP01033887 JAM69828.1 GDIQ01053392 JAN41345.1 GDIP01127067 JAL76647.1 GDIP01090770 JAM12945.1 GDIQ01196102 GDIQ01096169 JAL55557.1 GDIQ01123210 JAL28516.1 GFAH01000363 JAV48026.1
Proteomes
UP000008792
UP000192223
UP000007819
UP000036403
UP000075809
UP000078542
+ More
UP000242457 UP000005203 UP000005205 UP000078541 UP000078492 UP000078540 UP000279307 UP000053825 UP000075840 UP000075903 UP000053268 UP000053105 UP000075886 UP000283053 UP000009192 UP000095301 UP000007801 UP000095300 UP000215335 UP000007798 UP000092553 UP000192221 UP000009046 UP000008711 UP000000803 UP000002282 UP000001819 UP000268350 UP000235965 UP000000673 UP000007266 UP000076858
UP000242457 UP000005203 UP000005205 UP000078541 UP000078492 UP000078540 UP000279307 UP000053825 UP000075840 UP000075903 UP000053268 UP000053105 UP000075886 UP000283053 UP000009192 UP000095301 UP000007801 UP000095300 UP000215335 UP000007798 UP000092553 UP000192221 UP000009046 UP000008711 UP000000803 UP000002282 UP000001819 UP000268350 UP000235965 UP000000673 UP000007266 UP000076858
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
A0A1E1WTX5
B4LL10
A0A0Q9WGS5
A0A1W4XRS5
A0A1W4XQX5
A0A1W4XGR0
+ More
A0A2H8TR98 J9JUV1 A0A348G639 A0A0J7L5I2 A0A151X7M5 A0A195CBE9 A0A2A3E894 A0A088AJL9 A0A0C9R8B0 A0A158NA72 A0A195FR87 A0A195DMW3 A0A195BKD8 A0A3L8D615 A0A0L7QUX8 A0A1Q3FU39 A0A1I8JT87 A0A1B6BZK7 A0A2M4B8B1 A0A2M3ZY98 A0A2M3YXA0 A0A2M3YXA1 A0A1B6CXQ7 A0A182VCA9 A0A194QAS5 A0A0N0U341 A0A182QX48 A0A3S2NF93 W8AZY5 A0A0A1XSI3 A0A0K8WES0 A0A0K8TZ85 A0A1S4EYP3 A0A034W2L7 W4VRK7 V5JDF9 A0A0Q9X852 A0A1I8MRB0 T1PEM7 A0A0P9BZA8 A0A1I8NSL1 A0A0Q9X8Z6 A0A1L8E5H3 A0A0Q9XKV7 A0A1L8E5N4 A0A232EVU4 A0A0Q9WPB9 A0A0Q9WXH9 B4MJY8 A0A0M4EI37 A0A1W4V0I6 E0VNT1 A0A0Q5W3C6 A0A0J9RM65 Q59E58 A0A0R1DTW1 Q59E59 A0A1W4ULZ8 A0A0B4K7Q4 A0A0B4JD95 A0A0J9RL63 A0A0B4JD57 A0A224XCL1 A0A0J9RKL7 A0A0V0G429 A0A069DXW8 A0A146LQ23 A0A0R3NL29 A0A146MEA6 A0A146LT94 A0A1W4XRY8 A0A3B0J4P3 A0A2J7QWP2 A0A1W7RA51 Q99323-2 Q99323-1 W5JE10 A0A139WCQ8 A0A1Y1LEQ6 A0A1Y1LAZ4 V5GNY8 A0A0P5J9N0 A0A164ZVZ4 A0A0P5VW45 A0A0P5KU35 A0A0P5IY09 A0A0N8DH95 A0A0P6F3T2 A0A0P5UQL1 A0A0P5X1M2 A0A0P5SAS7 A0A0P5PUF7 A0A1W7RA82
A0A2H8TR98 J9JUV1 A0A348G639 A0A0J7L5I2 A0A151X7M5 A0A195CBE9 A0A2A3E894 A0A088AJL9 A0A0C9R8B0 A0A158NA72 A0A195FR87 A0A195DMW3 A0A195BKD8 A0A3L8D615 A0A0L7QUX8 A0A1Q3FU39 A0A1I8JT87 A0A1B6BZK7 A0A2M4B8B1 A0A2M3ZY98 A0A2M3YXA0 A0A2M3YXA1 A0A1B6CXQ7 A0A182VCA9 A0A194QAS5 A0A0N0U341 A0A182QX48 A0A3S2NF93 W8AZY5 A0A0A1XSI3 A0A0K8WES0 A0A0K8TZ85 A0A1S4EYP3 A0A034W2L7 W4VRK7 V5JDF9 A0A0Q9X852 A0A1I8MRB0 T1PEM7 A0A0P9BZA8 A0A1I8NSL1 A0A0Q9X8Z6 A0A1L8E5H3 A0A0Q9XKV7 A0A1L8E5N4 A0A232EVU4 A0A0Q9WPB9 A0A0Q9WXH9 B4MJY8 A0A0M4EI37 A0A1W4V0I6 E0VNT1 A0A0Q5W3C6 A0A0J9RM65 Q59E58 A0A0R1DTW1 Q59E59 A0A1W4ULZ8 A0A0B4K7Q4 A0A0B4JD95 A0A0J9RL63 A0A0B4JD57 A0A224XCL1 A0A0J9RKL7 A0A0V0G429 A0A069DXW8 A0A146LQ23 A0A0R3NL29 A0A146MEA6 A0A146LT94 A0A1W4XRY8 A0A3B0J4P3 A0A2J7QWP2 A0A1W7RA51 Q99323-2 Q99323-1 W5JE10 A0A139WCQ8 A0A1Y1LEQ6 A0A1Y1LAZ4 V5GNY8 A0A0P5J9N0 A0A164ZVZ4 A0A0P5VW45 A0A0P5KU35 A0A0P5IY09 A0A0N8DH95 A0A0P6F3T2 A0A0P5UQL1 A0A0P5X1M2 A0A0P5SAS7 A0A0P5PUF7 A0A1W7RA82
PDB
1I84
E-value=0,
Score=3120
Ontologies
GO
GO:0005524
GO:0016459
GO:0051015
GO:0003774
GO:0003779
GO:0016021
GO:0016887
GO:0060289
GO:0035277
GO:0007496
GO:0007395
GO:0071260
GO:0016461
GO:0007297
GO:0008258
GO:0045214
GO:0042060
GO:0032027
GO:0007455
GO:0042802
GO:0035159
GO:0007391
GO:0005826
GO:1901739
GO:0000281
GO:0035317
GO:0032154
GO:0044291
GO:0007443
GO:0046663
GO:1903688
GO:0046664
GO:0007435
GO:0005829
GO:0030239
GO:0106037
GO:0045200
GO:0006936
GO:0032507
GO:0035017
GO:0042623
GO:0005929
GO:0060571
GO:0016318
GO:0007298
GO:0005856
GO:0005938
GO:0005516
GO:0016460
GO:0031036
GO:0051259
GO:0030018
GO:0070986
GO:0001736
GO:0031252
GO:0045179
GO:0005515
GO:0043547
GO:0005634
GO:0004672
GO:0030677
GO:0016155
GO:0003824
Topology
Subcellular location
Expression in neurons and scolopale cells is increased at the apical tips of cilia. With evidence from 7 publications.
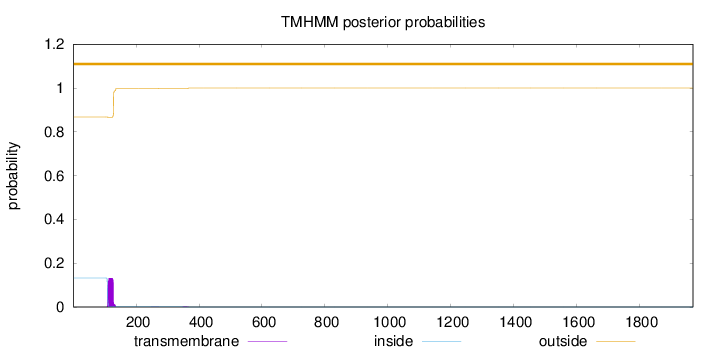
Length:
1970
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.64838000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.13218
outside
1 - 1970
Population Genetic Test Statistics
Pi
228.65259
Theta
169.580989
Tajima's D
0.825697
CLR
0.493938
CSRT
0.605319734013299
Interpretation
Uncertain
