Gene
KWMTBOMO10055 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005681
Annotation
PREDICTED:_L-allo-threonine_aldolase_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.498
Sequence
CDS
ATGGCACACGTAGTTGATTTACGTTCAGACACAGTTACAAAACCAACTAAAGCTATGAAACATGCAATGGTGAACTCTGCTTTGGGCGATGATGTTTACGGTGAAGATCCGACAGTAAACGCCTTGGAGAATAAAATAGCTGCACTGCTGGGCAAGCAGGCCGCTTTGTTCGTACCGTCTGGAACTATGTCTAATCTTATTGCCATTATGGTGCACTGTAACAAGCGTGGATCTGAAGCATTCGTCGGGGATCTCTCACACATTTATAAATATGAAGTTGGAGGAGCATCACATTTAGCAGGAGTGATGCTTTCGACCGTGGCCAATAAACCCGACGGCACTTTCGACCTGCAAGAGCTGGAGGATCGAATTAGGGGTTCGAATATACATGAACCAGTAACGGCTATGGTTGCCGTCGAAAATACCCATAATTACTGCGGAGGAAAGGTCCTGCCTTTGAAATTTCTTGACGACGTCGGCGCCTTGTGCAAGAAACACTCTCTGGCGCTGCACATGGACGGCGCGAGGCTGTTCAACGCGTGTGCGCATGCGCAGGAGGAGGCGGCGCGCGTGGTGCGGGACTGCGACTCCGTGTCCGTCTGCTTCAGCAAGGGACTGTCGGCCCCCGTCGGCTCGGCGCTCGTGGGGTCCTACCATTTCATCGAACAGGCCCGGCGCGCGCGCAAGATGCTGGGCGGCGGCATGCGGCAGGCCGGCGTGCTGGCGGCCGCGGCGCTGGTCGCGCTGCATGAGGTGGTGCCGCTGCTCGCCCTGGACCACAAGCGCGCCCAGGTGCTGGCCAAGGTTATCGAGGGTCTGTTCCTGAAGGCGTTCACCGTGGACGCGCACAACACGCACACGAATATCGTGCTCGTGCGCATCAGTGAGCACGCGCCGCTGACGGCCGACCAGCTCGTGCAGAGGCTGGCACAGGTCTGCCTCGCTGAAACCCAGGGAGACTGCAAGACGCCGAACGATGAAGGTGTGATCGTAAAAGCTACTGAGTTTAACTCGAAGACTATTCGATTCACTCTGCACCGCGAAATAACAGACGAAGACCTCTGGCTGGCCATCATGAAGATAACTTACGTTTTCAAAGAACTCAGTTCTAGTTTAGAGAACAAATAA
Protein
MAHVVDLRSDTVTKPTKAMKHAMVNSALGDDVYGEDPTVNALENKIAALLGKQAALFVPSGTMSNLIAIMVHCNKRGSEAFVGDLSHIYKYEVGGASHLAGVMLSTVANKPDGTFDLQELEDRIRGSNIHEPVTAMVAVENTHNYCGGKVLPLKFLDDVGALCKKHSLALHMDGARLFNACAHAQEEAARVVRDCDSVSVCFSKGLSAPVGSALVGSYHFIEQARRARKMLGGGMRQAGVLAAAALVALHEVVPLLALDHKRAQVLAKVIEGLFLKAFTVDAHNTHTNIVLVRISEHAPLTADQLVQRLAQVCLAETQGDCKTPNDEGVIVKATEFNSKTIRFTLHREITDEDLWLAIMKITYVFKELSSSLENK
Summary
Uniprot
H9J839
A0A2A4K8C2
A0A212ETH9
A0A0N1IDH2
A0A2H1WYH4
A0A3B0JGE4
+ More
Q178Q0 Q29BP8 A0A1S4FBQ3 B4GP99 A0A182GA83 B4JIB5 A0A182QXQ8 B3LY76 A0A2M4AVB1 B0WY29 A0A182W2Y6 A0A1Q3FQC8 A0A1W4V4S6 A0A0M4EN14 B4R218 A0A158N7L9 A0A182JFD5 Q9VCK6 A0A182NCJ9 A0A182F8D6 B3P8H0 B4N7A9 B4M0S3 B4HFV5 B4PNU9 B4KA71 U5ERL8 B4NJL6 A0A182YG69 A0A182WVG4 A0A182M6F0 A0A182I943 Q7QAI2 A0A182VCA1 A0A182PTS6 D3TMW4 A0A2J7QWQ6 A0A2J7QWS1 A0A182RZD2 T1PA97 A0A182TMA2 A0A1I8NCK1 A0A023ERH0 W8BMI9 A0A182K083 A0A1B0CA38 A0A0L0CQ19 A0A1A9WPR7 A0A034W4F4 A0A1L8E2X8 A0A336KNE6 A0A0A1WIW1 A0A0A1WKE7 A0A1L8E2G4 A0A1J1HQL3 A0A336KLE6 A0A1I8PWV6 A0A067QWE8 A0A182KXX1 A0A1B0DKN9 A0A1W4WFY4 E0VST6 A0A1Y1LAX9 A0A336KI28 A0A1Y1LLZ5 V5I9H7 A0A1Y1LM48 A0A1Y1LKG7 V5G655 A0A0P4W0E8 A0A3R7MTS9 A0A2L2Y5U3 A0A1W0XEX0 A0A2L2Y599 A0A1Y1M4H2 A0A1Y1M6K7 A0A1Y1M8A5 J3JUW4 J9JQ71 A0A1D1UT23 A0A2L2Y5G0 A0A226F636 A0A1B6EFI5 A0A2J7QWN2 A0A139WD87 E2B8B5 A0A1A9ZZA2 W5JHC8 A0A1A9YT50 A0A0V0G668 A0A1B0AKD1 A0A224XIE6 W5N4Z9 W5N4Z5
Q178Q0 Q29BP8 A0A1S4FBQ3 B4GP99 A0A182GA83 B4JIB5 A0A182QXQ8 B3LY76 A0A2M4AVB1 B0WY29 A0A182W2Y6 A0A1Q3FQC8 A0A1W4V4S6 A0A0M4EN14 B4R218 A0A158N7L9 A0A182JFD5 Q9VCK6 A0A182NCJ9 A0A182F8D6 B3P8H0 B4N7A9 B4M0S3 B4HFV5 B4PNU9 B4KA71 U5ERL8 B4NJL6 A0A182YG69 A0A182WVG4 A0A182M6F0 A0A182I943 Q7QAI2 A0A182VCA1 A0A182PTS6 D3TMW4 A0A2J7QWQ6 A0A2J7QWS1 A0A182RZD2 T1PA97 A0A182TMA2 A0A1I8NCK1 A0A023ERH0 W8BMI9 A0A182K083 A0A1B0CA38 A0A0L0CQ19 A0A1A9WPR7 A0A034W4F4 A0A1L8E2X8 A0A336KNE6 A0A0A1WIW1 A0A0A1WKE7 A0A1L8E2G4 A0A1J1HQL3 A0A336KLE6 A0A1I8PWV6 A0A067QWE8 A0A182KXX1 A0A1B0DKN9 A0A1W4WFY4 E0VST6 A0A1Y1LAX9 A0A336KI28 A0A1Y1LLZ5 V5I9H7 A0A1Y1LM48 A0A1Y1LKG7 V5G655 A0A0P4W0E8 A0A3R7MTS9 A0A2L2Y5U3 A0A1W0XEX0 A0A2L2Y599 A0A1Y1M4H2 A0A1Y1M6K7 A0A1Y1M8A5 J3JUW4 J9JQ71 A0A1D1UT23 A0A2L2Y5G0 A0A226F636 A0A1B6EFI5 A0A2J7QWN2 A0A139WD87 E2B8B5 A0A1A9ZZA2 W5JHC8 A0A1A9YT50 A0A0V0G668 A0A1B0AKD1 A0A224XIE6 W5N4Z9 W5N4Z5
Pubmed
19121390
22118469
26354079
17510324
15632085
17994087
+ More
26483478 20920257 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304 25244985 12364791 14747013 17210077 20353571 25315136 24945155 24495485 26108605 25348373 25830018 24845553 20966253 20566863 28004739 26561354 22516182 23537049 27649274 18362917 19820115 20798317
26483478 20920257 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304 25244985 12364791 14747013 17210077 20353571 25315136 24945155 24495485 26108605 25348373 25830018 24845553 20966253 20566863 28004739 26561354 22516182 23537049 27649274 18362917 19820115 20798317
EMBL
BABH01020389
NWSH01000033
PCG80477.1
AGBW02012580
OWR44790.1
KQ461156
+ More
KPJ08102.1 ODYU01011991 SOQ58052.1 OUUW01000005 SPP81225.1 CH477360 EAT42691.1 CM000070 EAL26949.2 CH479186 EDW38982.1 JXUM01050798 JXUM01050799 JXUM01050800 KQ561662 KXJ77888.1 CH916369 EDV92996.1 AXCN02001185 CH902617 EDV43980.1 GGFK01011331 MBW44652.1 DS232179 EDS36839.1 GFDL01005236 JAV29809.1 CP012526 ALC46940.1 CM000364 EDX14075.1 AE014297 AY051837 AAF56152.1 AAK93261.1 CH954182 EDV53994.1 CH964182 EDW80250.1 CH940650 EDW67365.1 KRF83263.1 CH480815 EDW43348.1 CM000160 EDW98159.1 CH933806 EDW16746.1 KRG02285.1 GANO01003674 JAB56197.1 CH964272 EDW83940.1 AXCM01004414 APCN01003173 AAAB01008888 EAA08948.4 EZ422766 ADD19042.1 NEVH01009422 PNF33005.1 PNF33004.1 KA645657 AFP60286.1 GAPW01002334 JAC11264.1 GAMC01004240 JAC02316.1 AJWK01003259 JRES01000065 KNC34450.1 GAKP01009942 JAC49010.1 GFDF01001152 JAV12932.1 UFQS01000513 UFQT01000513 SSX04513.1 SSX24876.1 GBXI01016004 JAC98287.1 GBXI01015282 JAC99009.1 GFDF01001151 JAV12933.1 CVRI01000010 CRK88766.1 UFQS01000608 UFQT01000608 SSX05319.1 SSX25680.1 KK853286 KDR08943.1 AJVK01068063 DS235756 EEB16442.1 GEZM01064535 JAV68736.1 SSX04514.1 SSX24877.1 GEZM01051876 JAV74653.1 GALX01002915 JAB65551.1 GEZM01057947 JAV72077.1 GEZM01057948 JAV72076.1 GALX01002916 JAB65550.1 GDRN01082358 GDRN01082354 JAI61844.1 QCYY01000194 ROT85764.1 IAAA01000823 IAAA01000824 LAA03367.1 MTYJ01000001 OQV25975.1 IAAA01000821 IAAA01000822 LAA03362.1 GEZM01042840 JAV79550.1 GEZM01042843 JAV79546.1 GEZM01042842 JAV79547.1 APGK01058541 BT127029 KB741291 AEE61991.1 ENN70507.1 ABLF02029828 ABLF02029829 BDGG01000002 GAU92834.1 IAAA01000825 LAA03368.1 LNIX01000001 OXA64967.1 GEDC01028749 GEDC01012315 GEDC01000647 JAS08549.1 JAS24983.1 JAS36651.1 PNF33003.1 KQ971361 KYB25872.1 GL446286 EFN88117.1 ADMH02001252 ETN63466.1 GECL01002561 JAP03563.1 JXJN01022789 GFTR01005602 JAW10824.1 AHAT01000366
KPJ08102.1 ODYU01011991 SOQ58052.1 OUUW01000005 SPP81225.1 CH477360 EAT42691.1 CM000070 EAL26949.2 CH479186 EDW38982.1 JXUM01050798 JXUM01050799 JXUM01050800 KQ561662 KXJ77888.1 CH916369 EDV92996.1 AXCN02001185 CH902617 EDV43980.1 GGFK01011331 MBW44652.1 DS232179 EDS36839.1 GFDL01005236 JAV29809.1 CP012526 ALC46940.1 CM000364 EDX14075.1 AE014297 AY051837 AAF56152.1 AAK93261.1 CH954182 EDV53994.1 CH964182 EDW80250.1 CH940650 EDW67365.1 KRF83263.1 CH480815 EDW43348.1 CM000160 EDW98159.1 CH933806 EDW16746.1 KRG02285.1 GANO01003674 JAB56197.1 CH964272 EDW83940.1 AXCM01004414 APCN01003173 AAAB01008888 EAA08948.4 EZ422766 ADD19042.1 NEVH01009422 PNF33005.1 PNF33004.1 KA645657 AFP60286.1 GAPW01002334 JAC11264.1 GAMC01004240 JAC02316.1 AJWK01003259 JRES01000065 KNC34450.1 GAKP01009942 JAC49010.1 GFDF01001152 JAV12932.1 UFQS01000513 UFQT01000513 SSX04513.1 SSX24876.1 GBXI01016004 JAC98287.1 GBXI01015282 JAC99009.1 GFDF01001151 JAV12933.1 CVRI01000010 CRK88766.1 UFQS01000608 UFQT01000608 SSX05319.1 SSX25680.1 KK853286 KDR08943.1 AJVK01068063 DS235756 EEB16442.1 GEZM01064535 JAV68736.1 SSX04514.1 SSX24877.1 GEZM01051876 JAV74653.1 GALX01002915 JAB65551.1 GEZM01057947 JAV72077.1 GEZM01057948 JAV72076.1 GALX01002916 JAB65550.1 GDRN01082358 GDRN01082354 JAI61844.1 QCYY01000194 ROT85764.1 IAAA01000823 IAAA01000824 LAA03367.1 MTYJ01000001 OQV25975.1 IAAA01000821 IAAA01000822 LAA03362.1 GEZM01042840 JAV79550.1 GEZM01042843 JAV79546.1 GEZM01042842 JAV79547.1 APGK01058541 BT127029 KB741291 AEE61991.1 ENN70507.1 ABLF02029828 ABLF02029829 BDGG01000002 GAU92834.1 IAAA01000825 LAA03368.1 LNIX01000001 OXA64967.1 GEDC01028749 GEDC01012315 GEDC01000647 JAS08549.1 JAS24983.1 JAS36651.1 PNF33003.1 KQ971361 KYB25872.1 GL446286 EFN88117.1 ADMH02001252 ETN63466.1 GECL01002561 JAP03563.1 JXJN01022789 GFTR01005602 JAW10824.1 AHAT01000366
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000268350
UP000008820
+ More
UP000001819 UP000008744 UP000069940 UP000249989 UP000001070 UP000075886 UP000007801 UP000002320 UP000075920 UP000192221 UP000092553 UP000000304 UP000075880 UP000000803 UP000075884 UP000069272 UP000008711 UP000007798 UP000008792 UP000001292 UP000002282 UP000009192 UP000076408 UP000076407 UP000075883 UP000075840 UP000007062 UP000075903 UP000075885 UP000235965 UP000075900 UP000075902 UP000095301 UP000075881 UP000092461 UP000037069 UP000091820 UP000183832 UP000095300 UP000027135 UP000075882 UP000092462 UP000192223 UP000009046 UP000283509 UP000019118 UP000007819 UP000186922 UP000198287 UP000007266 UP000008237 UP000092445 UP000000673 UP000092443 UP000092460 UP000018468
UP000001819 UP000008744 UP000069940 UP000249989 UP000001070 UP000075886 UP000007801 UP000002320 UP000075920 UP000192221 UP000092553 UP000000304 UP000075880 UP000000803 UP000075884 UP000069272 UP000008711 UP000007798 UP000008792 UP000001292 UP000002282 UP000009192 UP000076408 UP000076407 UP000075883 UP000075840 UP000007062 UP000075903 UP000075885 UP000235965 UP000075900 UP000075902 UP000095301 UP000075881 UP000092461 UP000037069 UP000091820 UP000183832 UP000095300 UP000027135 UP000075882 UP000092462 UP000192223 UP000009046 UP000283509 UP000019118 UP000007819 UP000186922 UP000198287 UP000007266 UP000008237 UP000092445 UP000000673 UP000092443 UP000092460 UP000018468
Interpro
Gene 3D
ProteinModelPortal
H9J839
A0A2A4K8C2
A0A212ETH9
A0A0N1IDH2
A0A2H1WYH4
A0A3B0JGE4
+ More
Q178Q0 Q29BP8 A0A1S4FBQ3 B4GP99 A0A182GA83 B4JIB5 A0A182QXQ8 B3LY76 A0A2M4AVB1 B0WY29 A0A182W2Y6 A0A1Q3FQC8 A0A1W4V4S6 A0A0M4EN14 B4R218 A0A158N7L9 A0A182JFD5 Q9VCK6 A0A182NCJ9 A0A182F8D6 B3P8H0 B4N7A9 B4M0S3 B4HFV5 B4PNU9 B4KA71 U5ERL8 B4NJL6 A0A182YG69 A0A182WVG4 A0A182M6F0 A0A182I943 Q7QAI2 A0A182VCA1 A0A182PTS6 D3TMW4 A0A2J7QWQ6 A0A2J7QWS1 A0A182RZD2 T1PA97 A0A182TMA2 A0A1I8NCK1 A0A023ERH0 W8BMI9 A0A182K083 A0A1B0CA38 A0A0L0CQ19 A0A1A9WPR7 A0A034W4F4 A0A1L8E2X8 A0A336KNE6 A0A0A1WIW1 A0A0A1WKE7 A0A1L8E2G4 A0A1J1HQL3 A0A336KLE6 A0A1I8PWV6 A0A067QWE8 A0A182KXX1 A0A1B0DKN9 A0A1W4WFY4 E0VST6 A0A1Y1LAX9 A0A336KI28 A0A1Y1LLZ5 V5I9H7 A0A1Y1LM48 A0A1Y1LKG7 V5G655 A0A0P4W0E8 A0A3R7MTS9 A0A2L2Y5U3 A0A1W0XEX0 A0A2L2Y599 A0A1Y1M4H2 A0A1Y1M6K7 A0A1Y1M8A5 J3JUW4 J9JQ71 A0A1D1UT23 A0A2L2Y5G0 A0A226F636 A0A1B6EFI5 A0A2J7QWN2 A0A139WD87 E2B8B5 A0A1A9ZZA2 W5JHC8 A0A1A9YT50 A0A0V0G668 A0A1B0AKD1 A0A224XIE6 W5N4Z9 W5N4Z5
Q178Q0 Q29BP8 A0A1S4FBQ3 B4GP99 A0A182GA83 B4JIB5 A0A182QXQ8 B3LY76 A0A2M4AVB1 B0WY29 A0A182W2Y6 A0A1Q3FQC8 A0A1W4V4S6 A0A0M4EN14 B4R218 A0A158N7L9 A0A182JFD5 Q9VCK6 A0A182NCJ9 A0A182F8D6 B3P8H0 B4N7A9 B4M0S3 B4HFV5 B4PNU9 B4KA71 U5ERL8 B4NJL6 A0A182YG69 A0A182WVG4 A0A182M6F0 A0A182I943 Q7QAI2 A0A182VCA1 A0A182PTS6 D3TMW4 A0A2J7QWQ6 A0A2J7QWS1 A0A182RZD2 T1PA97 A0A182TMA2 A0A1I8NCK1 A0A023ERH0 W8BMI9 A0A182K083 A0A1B0CA38 A0A0L0CQ19 A0A1A9WPR7 A0A034W4F4 A0A1L8E2X8 A0A336KNE6 A0A0A1WIW1 A0A0A1WKE7 A0A1L8E2G4 A0A1J1HQL3 A0A336KLE6 A0A1I8PWV6 A0A067QWE8 A0A182KXX1 A0A1B0DKN9 A0A1W4WFY4 E0VST6 A0A1Y1LAX9 A0A336KI28 A0A1Y1LLZ5 V5I9H7 A0A1Y1LM48 A0A1Y1LKG7 V5G655 A0A0P4W0E8 A0A3R7MTS9 A0A2L2Y5U3 A0A1W0XEX0 A0A2L2Y599 A0A1Y1M4H2 A0A1Y1M6K7 A0A1Y1M8A5 J3JUW4 J9JQ71 A0A1D1UT23 A0A2L2Y5G0 A0A226F636 A0A1B6EFI5 A0A2J7QWN2 A0A139WD87 E2B8B5 A0A1A9ZZA2 W5JHC8 A0A1A9YT50 A0A0V0G668 A0A1B0AKD1 A0A224XIE6 W5N4Z9 W5N4Z5
PDB
3WGB
E-value=9.59907e-70,
Score=669
Ontologies
PATHWAY
GO
PANTHER
Topology
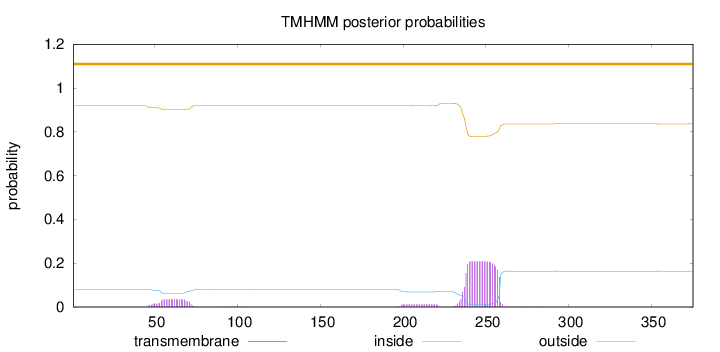
Length:
375
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.44758
Exp number, first 60 AAs:
0.34334
Total prob of N-in:
0.08061
outside
1 - 375
Population Genetic Test Statistics
Pi
168.86704
Theta
145.151704
Tajima's D
-1.01372
CLR
0.963152
CSRT
0.141042947852607
Interpretation
Uncertain
