Gene
KWMTBOMO10051
Pre Gene Modal
BGIBMGA005683
Annotation
Transmembrane_channel-like_protein_5_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.921
Sequence
CDS
ATGAAGTTGTTTCGAGGTATTGTCTGTCTACGGGCTGATGATGTTGTAATCCATAGATTGGAGTACTACACGCGTCGTACGGCGGTCTACGTGACGCTGGCCAGGACTTGGCTGTTGGACATGGGCGCGCTGATGCTTCTGCTGGTGTACTGGTCCCGGTCGAACACTGACTGCTGGGAGACGCGTTTCGGTCAGGAAGCGTACAGGCTGGTCCTGCTGGACGCCGTGGTCTCGCTGCTGGTGTTGCCAGCCATTGAGTTTATTAGAGCTTTGATATATAAGTTAAACCCGGACTCGTCGGCGCCGGAGTTCAACATAGCGTATAACTCGCTCACGTTGATTTACAACCAGGGGGTGCTCTGGTTCGGTCTGCTGTTCTCGCCCCTGCTCGTGGTGGCAGTCACCATCAAGTTCTTTATCTTGTTCTACGTGAAGAGAGAATGCGCTCTGAGGGCATGCCAGCCGGCTAGAAAGGTGTGGCGCGCGGCGCAGACGCAGACGGTGCTGTACGTGCTGGTCACGCTCTCGCTCTTCGCCACGCTGTTCGGGCTCGGCTCGCTCTTCTTCAGGAGTTCATCGAAGGTCTGCGGCCCGTTCCGTGGCTACGAGACGGTGTACAGCGTGCTCAGCGAAGGAGCCCTGAGGCTGTCGGAGCACCCCGCCGCCGCCACCATACTCGCCTTCTTCGGCAGGCCCGGACCTCTGGCCTTCATACTGCTTTTTCTCTGCGTGTCGGTGTACTACATGCGGGCGCGCGCCCTCGCGCACTCCTCCATGGTGGTCATATTGCGTCACATGCTAGTACTGCAGGCGAAGGACAAGGACTTCCTGCTCAACGCCATCGCTAAAGTCTCCAACGGGGAGTGGTCGTACAGCCCGAAGGCGGCCGAGGGTCCGGACAGCCACACGTGGAAGTACATTAGGGACGTCAGGAAGCCATCGAACTCTGGCTTCCATTTTGACGCATCGAGGCTCAGTCATTCGTTCGGGGACCGACCTCGGTCGCGCGTCAAGGAATACCGGCCTTATTCGCATTTGACCGAGAGGTCGAAATCGAATGACGGCGACACCGACAGCTCGTTTAGCTGGCAAGGCTCTAGCAGTCGTCTGGCACAAACCGAAGACGACAAGCGGTGGCCTTAG
Protein
MKLFRGIVCLRADDVVIHRLEYYTRRTAVYVTLARTWLLDMGALMLLLVYWSRSNTDCWETRFGQEAYRLVLLDAVVSLLVLPAIEFIRALIYKLNPDSSAPEFNIAYNSLTLIYNQGVLWFGLLFSPLLVVAVTIKFFILFYVKRECALRACQPARKVWRAAQTQTVLYVLVTLSLFATLFGLGSLFFRSSSKVCGPFRGYETVYSVLSEGALRLSEHPAAATILAFFGRPGPLAFILLFLCVSVYYMRARALAHSSMVVILRHMLVLQAKDKDFLLNAIAKVSNGEWSYSPKAAEGPDSHTWKYIRDVRKPSNSGFHFDASRLSHSFGDRPRSRVKEYRPYSHLTERSKSNDGDTDSSFSWQGSSSRLAQTEDDKRWP
Summary
Uniprot
A0A2A4K8Z1
A0A2H1VSY0
A0A3S2NV53
A0A0N1IPT7
A0A194QAR7
A0A2J7QE88
+ More
A0A067RJ61 A0A1S4FMK8 Q16VL2 V5GL48 A0A023EUS1 A0A182H2F3 A0A1Q3FCV5 B0X6W4 A0A1B6C112 A0A1B6DMG2 A0A1Y1K0S4 A0A1W4WJ38 A0A1W4WJ20 A0A1B6GBE1 A0A1J1I8S3 A0A0K8VHL7 A0A336LSF5 A0A0C9RPE8 U4TWT5 N6TQJ7 A0A0K8VN84 A0A0A1X885 A0A034VS87 A0A0C9R2I4 A0A139WEB7 E2AP44 E2BAB1 A0A232F3N7 W8BII4 K7IYE9 A0A2M3ZLV2 A0A1L8DDF2 A0A2M3ZLS5 A0A2M4A3F4 A0A1L8DDI1 A0A1L8DEZ3 A0A1L8DF41 A0A026VT44 A0A182J950 W5JPD2 A0A195DN52 A0A1E1XQS6 A0A182SE38 A0A1E1X6D3 F4W4F6 A0A151WYX9 A0A195BHD7 A0A182K6H9 A0A131XN56 A0A182N108 A0A182RV51 A0A182W4Q7 A0A158P373 A0A210QNW6 A0A195F0E0 A0A182QU36 A0A131YW06 A0A084VAB2 A0A224Z8K8 A0A195CD28 A0A182Y7H2 A0A182MS89 A0A182XLP5 V4A250 A0A182VLE1 A0A182IA56 A0A182TSQ3 A0A2R5LEM3 Q7Z014 A0A182PQ61 F7ITZ5 A0A1S4G9C4 A0A0P4W0Z3 A0A0P4W4G5 A0A0P4VVR6 A0A2C9M3S8 A0A293M4R7 A0A293LIL5
A0A067RJ61 A0A1S4FMK8 Q16VL2 V5GL48 A0A023EUS1 A0A182H2F3 A0A1Q3FCV5 B0X6W4 A0A1B6C112 A0A1B6DMG2 A0A1Y1K0S4 A0A1W4WJ38 A0A1W4WJ20 A0A1B6GBE1 A0A1J1I8S3 A0A0K8VHL7 A0A336LSF5 A0A0C9RPE8 U4TWT5 N6TQJ7 A0A0K8VN84 A0A0A1X885 A0A034VS87 A0A0C9R2I4 A0A139WEB7 E2AP44 E2BAB1 A0A232F3N7 W8BII4 K7IYE9 A0A2M3ZLV2 A0A1L8DDF2 A0A2M3ZLS5 A0A2M4A3F4 A0A1L8DDI1 A0A1L8DEZ3 A0A1L8DF41 A0A026VT44 A0A182J950 W5JPD2 A0A195DN52 A0A1E1XQS6 A0A182SE38 A0A1E1X6D3 F4W4F6 A0A151WYX9 A0A195BHD7 A0A182K6H9 A0A131XN56 A0A182N108 A0A182RV51 A0A182W4Q7 A0A158P373 A0A210QNW6 A0A195F0E0 A0A182QU36 A0A131YW06 A0A084VAB2 A0A224Z8K8 A0A195CD28 A0A182Y7H2 A0A182MS89 A0A182XLP5 V4A250 A0A182VLE1 A0A182IA56 A0A182TSQ3 A0A2R5LEM3 Q7Z014 A0A182PQ61 F7ITZ5 A0A1S4G9C4 A0A0P4W0Z3 A0A0P4W4G5 A0A0P4VVR6 A0A2C9M3S8 A0A293M4R7 A0A293LIL5
Pubmed
26354079
24845553
17510324
24945155
26483478
28004739
+ More
23537049 25830018 25348373 18362917 19820115 20798317 28648823 24495485 20075255 24508170 30249741 20920257 23761445 29209593 28503490 21719571 28049606 21347285 28812685 26830274 24438588 28797301 25244985 23254933 12812529 12364791 15562597
23537049 25830018 25348373 18362917 19820115 20798317 28648823 24495485 20075255 24508170 30249741 20920257 23761445 29209593 28503490 21719571 28049606 21347285 28812685 26830274 24438588 28797301 25244985 23254933 12812529 12364791 15562597
EMBL
NWSH01000033
PCG80466.1
ODYU01004264
SOQ43951.1
RSAL01000062
RVE49557.1
+ More
KQ460044 KPJ18331.1 KQ459232 KPJ02562.1 NEVH01015318 PNF26902.1 KK852652 KDR19382.1 CH477589 EAT38584.1 GALX01003647 JAB64819.1 GAPW01000418 JAC13180.1 JXUM01024284 KQ560685 KXJ81282.1 GFDL01009631 JAV25414.1 DS232427 EDS41635.1 GEDC01030339 JAS06959.1 GEDC01010450 JAS26848.1 GEZM01098847 JAV53720.1 GECZ01010018 JAS59751.1 CVRI01000044 CRK96627.1 GDHF01013961 JAI38353.1 UFQT01000140 SSX20755.1 GBYB01010305 JAG80072.1 KB631579 ERL84433.1 APGK01058707 KB741291 ENN70561.1 GDHF01011973 JAI40341.1 GBXI01006995 JAD07297.1 GAKP01013633 JAC45319.1 GBYB01010304 JAG80071.1 KQ971357 KYB26181.1 GL441452 EFN64780.1 GL446696 EFN87366.1 NNAY01001116 OXU25088.1 GAMC01008018 JAB98537.1 GGFM01008734 MBW29485.1 GFDF01009585 JAV04499.1 GGFM01008735 MBW29486.1 GGFK01002023 MBW35344.1 GFDF01009586 JAV04498.1 GFDF01009129 JAV04955.1 GFDF01009130 JAV04954.1 KK108051 QOIP01000001 EZA46953.1 RLU26903.1 AXCP01007156 ADMH02000938 ETN64624.1 KQ980713 KYN14256.1 GFAA01002133 JAU01302.1 GFAC01004577 JAT94611.1 GL887532 EGI70851.1 KQ982649 KYQ53102.1 KQ976467 KYM84184.1 GEFH01001031 JAP67550.1 ADTU01007769 NEDP02002607 OWF50434.1 KQ981905 KYN33584.1 AXCN02001393 GEDV01005857 JAP82700.1 ATLV01003054 KE524109 KFB34906.1 GFPF01012775 MAA23921.1 KQ977935 KYM98707.1 AXCM01005583 KB202619 ESO89005.1 APCN01001124 GGLE01003845 MBY07971.1 AY263176 AAP78791.1 AAAB01008846 EAA06337.4 GDRN01092930 GDRN01092929 JAI60005.1 GDRN01092925 JAI60007.1 GDRN01092927 JAI60006.1 GFWV01010309 MAA35038.1 GFWV01002969 MAA27699.1
KQ460044 KPJ18331.1 KQ459232 KPJ02562.1 NEVH01015318 PNF26902.1 KK852652 KDR19382.1 CH477589 EAT38584.1 GALX01003647 JAB64819.1 GAPW01000418 JAC13180.1 JXUM01024284 KQ560685 KXJ81282.1 GFDL01009631 JAV25414.1 DS232427 EDS41635.1 GEDC01030339 JAS06959.1 GEDC01010450 JAS26848.1 GEZM01098847 JAV53720.1 GECZ01010018 JAS59751.1 CVRI01000044 CRK96627.1 GDHF01013961 JAI38353.1 UFQT01000140 SSX20755.1 GBYB01010305 JAG80072.1 KB631579 ERL84433.1 APGK01058707 KB741291 ENN70561.1 GDHF01011973 JAI40341.1 GBXI01006995 JAD07297.1 GAKP01013633 JAC45319.1 GBYB01010304 JAG80071.1 KQ971357 KYB26181.1 GL441452 EFN64780.1 GL446696 EFN87366.1 NNAY01001116 OXU25088.1 GAMC01008018 JAB98537.1 GGFM01008734 MBW29485.1 GFDF01009585 JAV04499.1 GGFM01008735 MBW29486.1 GGFK01002023 MBW35344.1 GFDF01009586 JAV04498.1 GFDF01009129 JAV04955.1 GFDF01009130 JAV04954.1 KK108051 QOIP01000001 EZA46953.1 RLU26903.1 AXCP01007156 ADMH02000938 ETN64624.1 KQ980713 KYN14256.1 GFAA01002133 JAU01302.1 GFAC01004577 JAT94611.1 GL887532 EGI70851.1 KQ982649 KYQ53102.1 KQ976467 KYM84184.1 GEFH01001031 JAP67550.1 ADTU01007769 NEDP02002607 OWF50434.1 KQ981905 KYN33584.1 AXCN02001393 GEDV01005857 JAP82700.1 ATLV01003054 KE524109 KFB34906.1 GFPF01012775 MAA23921.1 KQ977935 KYM98707.1 AXCM01005583 KB202619 ESO89005.1 APCN01001124 GGLE01003845 MBY07971.1 AY263176 AAP78791.1 AAAB01008846 EAA06337.4 GDRN01092930 GDRN01092929 JAI60005.1 GDRN01092925 JAI60007.1 GDRN01092927 JAI60006.1 GFWV01010309 MAA35038.1 GFWV01002969 MAA27699.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000235965
UP000027135
+ More
UP000008820 UP000069940 UP000249989 UP000002320 UP000192223 UP000183832 UP000030742 UP000019118 UP000007266 UP000000311 UP000008237 UP000215335 UP000002358 UP000053097 UP000279307 UP000075880 UP000000673 UP000078492 UP000075901 UP000007755 UP000075809 UP000078540 UP000075881 UP000075884 UP000075900 UP000075920 UP000005205 UP000242188 UP000078541 UP000075886 UP000030765 UP000078542 UP000076408 UP000075883 UP000076407 UP000030746 UP000075903 UP000075840 UP000075902 UP000075885 UP000007062 UP000076420
UP000008820 UP000069940 UP000249989 UP000002320 UP000192223 UP000183832 UP000030742 UP000019118 UP000007266 UP000000311 UP000008237 UP000215335 UP000002358 UP000053097 UP000279307 UP000075880 UP000000673 UP000078492 UP000075901 UP000007755 UP000075809 UP000078540 UP000075881 UP000075884 UP000075900 UP000075920 UP000005205 UP000242188 UP000078541 UP000075886 UP000030765 UP000078542 UP000076408 UP000075883 UP000076407 UP000030746 UP000075903 UP000075840 UP000075902 UP000075885 UP000007062 UP000076420
Pfam
PF07810 TMC
ProteinModelPortal
A0A2A4K8Z1
A0A2H1VSY0
A0A3S2NV53
A0A0N1IPT7
A0A194QAR7
A0A2J7QE88
+ More
A0A067RJ61 A0A1S4FMK8 Q16VL2 V5GL48 A0A023EUS1 A0A182H2F3 A0A1Q3FCV5 B0X6W4 A0A1B6C112 A0A1B6DMG2 A0A1Y1K0S4 A0A1W4WJ38 A0A1W4WJ20 A0A1B6GBE1 A0A1J1I8S3 A0A0K8VHL7 A0A336LSF5 A0A0C9RPE8 U4TWT5 N6TQJ7 A0A0K8VN84 A0A0A1X885 A0A034VS87 A0A0C9R2I4 A0A139WEB7 E2AP44 E2BAB1 A0A232F3N7 W8BII4 K7IYE9 A0A2M3ZLV2 A0A1L8DDF2 A0A2M3ZLS5 A0A2M4A3F4 A0A1L8DDI1 A0A1L8DEZ3 A0A1L8DF41 A0A026VT44 A0A182J950 W5JPD2 A0A195DN52 A0A1E1XQS6 A0A182SE38 A0A1E1X6D3 F4W4F6 A0A151WYX9 A0A195BHD7 A0A182K6H9 A0A131XN56 A0A182N108 A0A182RV51 A0A182W4Q7 A0A158P373 A0A210QNW6 A0A195F0E0 A0A182QU36 A0A131YW06 A0A084VAB2 A0A224Z8K8 A0A195CD28 A0A182Y7H2 A0A182MS89 A0A182XLP5 V4A250 A0A182VLE1 A0A182IA56 A0A182TSQ3 A0A2R5LEM3 Q7Z014 A0A182PQ61 F7ITZ5 A0A1S4G9C4 A0A0P4W0Z3 A0A0P4W4G5 A0A0P4VVR6 A0A2C9M3S8 A0A293M4R7 A0A293LIL5
A0A067RJ61 A0A1S4FMK8 Q16VL2 V5GL48 A0A023EUS1 A0A182H2F3 A0A1Q3FCV5 B0X6W4 A0A1B6C112 A0A1B6DMG2 A0A1Y1K0S4 A0A1W4WJ38 A0A1W4WJ20 A0A1B6GBE1 A0A1J1I8S3 A0A0K8VHL7 A0A336LSF5 A0A0C9RPE8 U4TWT5 N6TQJ7 A0A0K8VN84 A0A0A1X885 A0A034VS87 A0A0C9R2I4 A0A139WEB7 E2AP44 E2BAB1 A0A232F3N7 W8BII4 K7IYE9 A0A2M3ZLV2 A0A1L8DDF2 A0A2M3ZLS5 A0A2M4A3F4 A0A1L8DDI1 A0A1L8DEZ3 A0A1L8DF41 A0A026VT44 A0A182J950 W5JPD2 A0A195DN52 A0A1E1XQS6 A0A182SE38 A0A1E1X6D3 F4W4F6 A0A151WYX9 A0A195BHD7 A0A182K6H9 A0A131XN56 A0A182N108 A0A182RV51 A0A182W4Q7 A0A158P373 A0A210QNW6 A0A195F0E0 A0A182QU36 A0A131YW06 A0A084VAB2 A0A224Z8K8 A0A195CD28 A0A182Y7H2 A0A182MS89 A0A182XLP5 V4A250 A0A182VLE1 A0A182IA56 A0A182TSQ3 A0A2R5LEM3 Q7Z014 A0A182PQ61 F7ITZ5 A0A1S4G9C4 A0A0P4W0Z3 A0A0P4W4G5 A0A0P4VVR6 A0A2C9M3S8 A0A293M4R7 A0A293LIL5
Ontologies
KEGG
GO
PANTHER
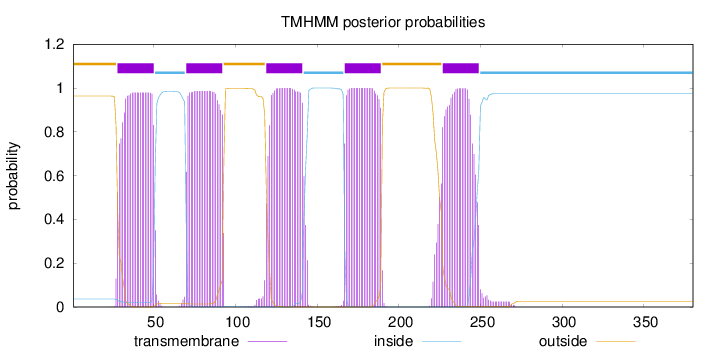
Topology
Length:
380
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
113.06998
Exp number, first 60 AAs:
21.7716
Total prob of N-in:
0.03672
POSSIBLE N-term signal
sequence
outside
1 - 27
TMhelix
28 - 50
inside
51 - 69
TMhelix
70 - 92
outside
93 - 118
TMhelix
119 - 141
inside
142 - 166
TMhelix
167 - 189
outside
190 - 226
TMhelix
227 - 249
inside
250 - 380
Population Genetic Test Statistics
Pi
203.437217
Theta
179.070566
Tajima's D
-1.851523
CLR
318.959474
CSRT
0.0243987800609969
Interpretation
Possibly Positive selection
