Pre Gene Modal
BGIBMGA005685
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_chaoptin-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.195 PlasmaMembrane Reliability : 1.841
Sequence
CDS
ATGAGGCGAGGTGCGATGCAGCAATGGTCAGCGTGCTGGATCCACGCGGCGCTCGCTGTGATGCTGTTGCCCGGAGTGAGGGCTCCAGCCCGTCCCTGTCCAAGGAACTCGCTCTGCTATTGTCGCGCGGACCACTTCGCCTGCAACTACGTTCCGTTCCATCGTTTCCCAGAAACAGATCCAGAAGTATGGCACGTGTCAGTGTCGCAAGCTCGCTTGGGTACCCTGGGTGAGGGTGCACTGGATGGGAGACGGCTGCGGACCCTGGTGCTGGTAGCTTCACATCTATACCACATAGAACCCGGAGCTTTTTCGTCAATGACGTCAACATTGGCGTCCTTGGACTTAGCCAACAATGAATTCACAGAAATCCCTATCGAAGCTCTTCGAGAATTAAAAGTGTTGAATTGGCTCAATTTGCAAAACAATTACATTAGCGAATTAAATTCGGATTTTGACTGGGGTTTCTTATCCGATTCTTTGAGCAGCCTATCATTAAGCAACAATCATCTGTATATCCTGAACGAAGGTTCATTATCGTCACTACATCAGCTGGCCCAGCTCGATCTTGACGGGAATCGATTACATGCCGTCAACCCGAATTCGCTGCCACAGTCATTGGCATTGCTTCGTCTGTCTGATAACATATTGACATACTTCCCATGTCTAACGTTGGCTGGGTTACCACGTCTACTACACCTTCACCTCAGAAATAACATACTAAGACCTTCCTTAAACCACACGTGCAGGGGTGAACGCACCAAAATAGAGTCATTAGATTTGAGTCACAATGAGCTGGATGATCGTTTTGTTCTCAGCTTCCAGCACAGATTACAGCTGAAGCAATTAATATTAGACTTTAATGATTTTACGGCCATTCCTCCTTTCGTTATGGACAGCGGTCGGTTAGAGAAATTATCAATTTCTTACAATAAATTAAGCTATGTGTCAGATGCGATGATCCATGCATTGAAAAGAGATTTGGAAAGATTGGAACTGGATTACAATGAAATAGCTATTCTATCAGGTAGTGTTAAGGAGATGGTGAGACTTAAGCATTTGTCGCTAGCTTACAATCACCTAGAAGAGATCAATGATCTTCCTCCAAATTTGTATTCTTTGTCATTGTCCGGGAATTTCCTGTTGACTTTTCCAGTGGGCTTAAGTACTTTAAGTCCTGCGAGTCTGGCGTATTTAGATCTCGGATATAATCAAATATCGGTCTTGTCCCCCGAATCCTTCGGTGTGTGGTCGGAAGCACTCGTAACTCTAAATCTCAAAGGGAATAGGATCACACAGCTCGTGGCGGGGTCCTTCCCTTTAACTTTACCCTTAAGAGAACTTGTACTCAGCTTCAATGACCTTTATTACGTAGATCCAAATGCTTTTGCAAATCTAACGATGTTGCAAGTCTTAGAACTATCTTCGACTTTATTCAGTGGAGAATTTCCAGTGTCTTCGTATTTCGATAATCTGACTTGGCTGAGTTTAGATAACAATAACGTTCATTACGTCTCGTCCGGGGACCTCCAGAACTTTCCCTCACTACAATATCTCAATTTAGAGTTCAATAAGATAACAGAGTTTCCTAGTGAAGTCACTCTAACGTCTGATCGATCGAACAGACTTAAAGAACTGAGACTATCATATAATTACATAAGCAAGATAAATTCGGAATTCCTCTCCGATTTAGTGGAGTTGCAAAGTTTAGATTTATCGTATAATCGCGTGTACAATATTAGCGAGAACGGTTTCTCGAATTTACACAATTTGGTTTATTTGAGCTTGTCCGGGAACGTTGTGGAGTTCATAGCGGAACGTGCGTTCCGACGCTTGCCCAAATTAGAGGCGTTAGACATGCACGAGAATAATTTACTCGTATTCTCTACGGACTGTTTCGAAAATGTTTCCAACGATGAAACAAACTTTTCGGTAAACGTTAGCCGAAATAGAATATCCGTGTTGACCGGCGGGCTGGCCGTTTCAATAAATACACTCGATCTATCCTTTAATTTTTTAGAGAGCGTCTCACGACATTTTTTTGACTCAATCGGACCAGCCTTGCGACAGTTAATTGTTTCGCACAACAGATTAATGCACCTCGATAACATTGCGTTTGGCTTCCTGCCATCTCTAGAAGTATTGATGCTACACGATAACAGTATAAGTGCTATTAAAAAGAAAGCCTTCGCTGAGATGTCGTCGCTTCAAATCCTGGATCTCTCACGTAATAAAATCAGCCAATTAGCTGTAGAGCAATTTCATAATATGCGACGCTTAAGGCATCTGCGCTTGGCGCGAAATGTCCTGCGATCTTTGCCACGAGATGTATTTAAAAACACCATTCTCGAACATTTAGACCTTAATGACAATCATATTGCAATATTTCCCAGTAGTGCTTTGGCTCAAGTCGGTTTCACTTTAAGAAGACTGGAACTAGCTCAAAATCACCTAGAATATCTCGACGCGGCGATGTTCCACGCTATCGCTTTTCTTCACGAGCTGACACTCGCTCGTAATGCTCTTACAGTACTTTCGGATAACACATTCGCTGGATTAACCAGACTGCGACAGTTAGACTTATCACATAATCCCGTTAAGACGAATTTCAAGGAACTGTTTCACAACCTTCCTCGACTGCGCAAGTTGATCCTCGCCGACGCTGGCCTTAAGAGTATACCGCATTTGCCACTTGCTAATCTAACTGAACTGGATCTGAGTGGTAACTTCATGTCATCTTTTCGGGAGCCTGAAGTGAGGCGATTGATGAATCTACGCGCGCTGAACTTGGCAAGAAATAAATTTACGTCATTGCAACCAGCCATGTGGGCGGCGTTACCGAAATTGTGTAACCTTGACGTATCACAGAACCCTATAGTCCGAGTGACTCGTGGATCATTCGAAGGTCTCGACCGCTTACTACATTTACGGATGCCACAGCTAAGGCAGTTAGAGGTGGTGGAGCCCCGAGCGTTCAGGCCATTAGTTTCTCTTCGGATTTTGGATTTAGAATCGCCCATTGGATCGGGTCGAGCAGAAGCATCCCTTGCCGATATTTCTCTATCAGTACCCACACTGGAATCTCTCTCTGTTTTGGTCAGAGAAAGTGTTCTCGATACCCAACTACATGGGCTAAGAGCACCGAAATTGCGGTCACTTGAGGTGCATGGAGTGGCGTTGCGTCGTGTCTCTGTGCATGCGTTTGTATCTTTGGGCCAGCAGCGTGCCTTGACTCTACGATTGCGACGAACTAGCATCACTGCACTCCCACCAGGTATTGTGAAACCCGTGGCCCGTGTCCCACACCTGGCACTGGACTTCAGTGCTAACCAGTTAGTTACCTTTGGTCCGGCGACACTCTACCCAAACTTGACTGGTTGGAACCGTCTCGCAACAAAACTCCTCCCAGGAGGCCTCATACTATCAGAGAATCCACTCCGTTGTGGTTGCTCTGTATCGTGGGTGGGGGCTTGGTTGCGCCGTTGGACAGCTGAGGTCGGAGGCTGGTCAAGAGACGGTCGTGATGCAGCACGACGTAGCGCGTGCATGATATCCGGGACACTAGTTCCACTCCTGTCTCTGGAGGCAGATGAGGCGGAGTGTCACGCCAGTGCACTCTCAAGTAGAGCATTAACACATCGTCCTCACACAATGAGATCATTTCAATGGATCGTGTTATTATATGCTCTGAGTTAG
Protein
MRRGAMQQWSACWIHAALAVMLLPGVRAPARPCPRNSLCYCRADHFACNYVPFHRFPETDPEVWHVSVSQARLGTLGEGALDGRRLRTLVLVASHLYHIEPGAFSSMTSTLASLDLANNEFTEIPIEALRELKVLNWLNLQNNYISELNSDFDWGFLSDSLSSLSLSNNHLYILNEGSLSSLHQLAQLDLDGNRLHAVNPNSLPQSLALLRLSDNILTYFPCLTLAGLPRLLHLHLRNNILRPSLNHTCRGERTKIESLDLSHNELDDRFVLSFQHRLQLKQLILDFNDFTAIPPFVMDSGRLEKLSISYNKLSYVSDAMIHALKRDLERLELDYNEIAILSGSVKEMVRLKHLSLAYNHLEEINDLPPNLYSLSLSGNFLLTFPVGLSTLSPASLAYLDLGYNQISVLSPESFGVWSEALVTLNLKGNRITQLVAGSFPLTLPLRELVLSFNDLYYVDPNAFANLTMLQVLELSSTLFSGEFPVSSYFDNLTWLSLDNNNVHYVSSGDLQNFPSLQYLNLEFNKITEFPSEVTLTSDRSNRLKELRLSYNYISKINSEFLSDLVELQSLDLSYNRVYNISENGFSNLHNLVYLSLSGNVVEFIAERAFRRLPKLEALDMHENNLLVFSTDCFENVSNDETNFSVNVSRNRISVLTGGLAVSINTLDLSFNFLESVSRHFFDSIGPALRQLIVSHNRLMHLDNIAFGFLPSLEVLMLHDNSISAIKKKAFAEMSSLQILDLSRNKISQLAVEQFHNMRRLRHLRLARNVLRSLPRDVFKNTILEHLDLNDNHIAIFPSSALAQVGFTLRRLELAQNHLEYLDAAMFHAIAFLHELTLARNALTVLSDNTFAGLTRLRQLDLSHNPVKTNFKELFHNLPRLRKLILADAGLKSIPHLPLANLTELDLSGNFMSSFREPEVRRLMNLRALNLARNKFTSLQPAMWAALPKLCNLDVSQNPIVRVTRGSFEGLDRLLHLRMPQLRQLEVVEPRAFRPLVSLRILDLESPIGSGRAEASLADISLSVPTLESLSVLVRESVLDTQLHGLRAPKLRSLEVHGVALRRVSVHAFVSLGQQRALTLRLRRTSITALPPGIVKPVARVPHLALDFSANQLVTFGPATLYPNLTGWNRLATKLLPGGLILSENPLRCGCSVSWVGAWLRRWTAEVGGWSRDGRDAARRSACMISGTLVPLLSLEADEAECHASALSSRALTHRPHTMRSFQWIVLLYALS
Summary
Uniprot
H9J843
A0A0N0PE55
A0A2A4JKM1
A0A2W1BDV4
A0A2H1W8H3
A0A212EKW8
+ More
A0A2A4JN70 A0A3S2LVH4 A0A194QH46 E0VKV3 A0A212ET41 A0A0L7LUG8 D6WHI0 A0A2J7PKP3 A0A2P8YPX1 A0A067QSJ1 A0A2H8TK33 A0A2S2R4K0 B4JAY0 A0A0R3NR54 B4GK90 Q29P78 B4LRW9 A0A1B6GDA1 A0A0Q9WG37 B3MM94 A0A0J9TNN6 B4NYD5 A0A1B0DDM6 A0A0N8NZ90 A0A0P8XG79 Q9VJQ0 A0A0J9R2M7 B4KLA8 A0A1W4VY78 B3N5P6 A0A0R1DN70 A0A0Q9XFX5 A0A0Q5W9N8 B4MZE7 A0A1J1IQY4 A0A0L0C4D4 B4HXM8 J9JTX0 A0A1I8M578 A0A0R3NQR3 Q7PWD3 A0A1A9WZG1 A0A1A9XY19 A0A0Q9WA29 A0A1I8QBR3 A0A1I8QBR1 A0A1B0BDC5 A0A1A9UH23 A0A3B0KMR9 A0A336MLY2 A0A1W4WPN6 A0A336MVI4 A0A1B0FRG7 A0A1A9Z997 Q17LV0 A0A1S4EYA8 A0A154PBK7 E2B3W6 B0W620 A0A2A3EMA2 A0A084VKG4 A0A088AAQ5 A0A182P6W4 A0A2S2NNB4 A0A182YFT1 A0A0M9A8L2 A0A182F265 A0A182PZJ5 A0A182I7L9 A0A182TZN0 A0A182R6L0 A0A026WGI9 A0A182WXQ5 A0A182NBF6 A0A182LMF4 A0A182IXJ5 A0A182VU11 A0A1S4GZU6 A0A1B6CFS9 W5JP87 A0A158NLR7 A0A195DB55 E2AH27 A0A182LWN1 A0A195C2N7 F4WXH7 A0A195AXS6 K7IWS3 A0A195F4W9 A0A151X857 A0A1B0CY43
A0A2A4JN70 A0A3S2LVH4 A0A194QH46 E0VKV3 A0A212ET41 A0A0L7LUG8 D6WHI0 A0A2J7PKP3 A0A2P8YPX1 A0A067QSJ1 A0A2H8TK33 A0A2S2R4K0 B4JAY0 A0A0R3NR54 B4GK90 Q29P78 B4LRW9 A0A1B6GDA1 A0A0Q9WG37 B3MM94 A0A0J9TNN6 B4NYD5 A0A1B0DDM6 A0A0N8NZ90 A0A0P8XG79 Q9VJQ0 A0A0J9R2M7 B4KLA8 A0A1W4VY78 B3N5P6 A0A0R1DN70 A0A0Q9XFX5 A0A0Q5W9N8 B4MZE7 A0A1J1IQY4 A0A0L0C4D4 B4HXM8 J9JTX0 A0A1I8M578 A0A0R3NQR3 Q7PWD3 A0A1A9WZG1 A0A1A9XY19 A0A0Q9WA29 A0A1I8QBR3 A0A1I8QBR1 A0A1B0BDC5 A0A1A9UH23 A0A3B0KMR9 A0A336MLY2 A0A1W4WPN6 A0A336MVI4 A0A1B0FRG7 A0A1A9Z997 Q17LV0 A0A1S4EYA8 A0A154PBK7 E2B3W6 B0W620 A0A2A3EMA2 A0A084VKG4 A0A088AAQ5 A0A182P6W4 A0A2S2NNB4 A0A182YFT1 A0A0M9A8L2 A0A182F265 A0A182PZJ5 A0A182I7L9 A0A182TZN0 A0A182R6L0 A0A026WGI9 A0A182WXQ5 A0A182NBF6 A0A182LMF4 A0A182IXJ5 A0A182VU11 A0A1S4GZU6 A0A1B6CFS9 W5JP87 A0A158NLR7 A0A195DB55 E2AH27 A0A182LWN1 A0A195C2N7 F4WXH7 A0A195AXS6 K7IWS3 A0A195F4W9 A0A151X857 A0A1B0CY43
Pubmed
19121390
26354079
28756777
22118469
20566863
26227816
+ More
18362917 19820115 29403074 24845553 17994087 15632085 23185243 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 25315136 12364791 17510324 20798317 24438588 25244985 24508170 30249741 20966253 20920257 23761445 21347285 21719571 20075255
18362917 19820115 29403074 24845553 17994087 15632085 23185243 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 25315136 12364791 17510324 20798317 24438588 25244985 24508170 30249741 20966253 20920257 23761445 21347285 21719571 20075255
EMBL
BABH01020400
KQ460044
KPJ18286.1
NWSH01001260
PCG71950.1
KZ150179
+ More
PZC72541.1 ODYU01006995 SOQ49347.1 AGBW02014188 OWR42121.1 NWSH01001061 PCG72850.1 RSAL01000168 RVE45242.1 KQ459232 KPJ02766.1 DS235252 EEB14009.1 AGBW02012643 OWR44663.1 JTDY01000063 KOB79092.1 KQ971321 EFA01333.2 NEVH01024535 PNF16900.1 PYGN01000441 PSN46305.1 KK853075 KDR11757.1 GFXV01002689 MBW14494.1 GGMS01015681 MBY84884.1 CH916368 EDW02850.1 CH379058 KRT03481.1 CH479184 EDW37056.1 EAL34415.3 KRT03478.1 KRT03479.1 CH940649 EDW63645.1 GECZ01009379 JAS60390.1 KRF81223.1 CH902620 EDV31854.2 CM002910 KMY90345.1 CM000157 EDW89771.2 AJVK01005457 AJVK01005458 AJVK01005459 AJVK01005460 KPU73660.1 KPU73659.1 AE014134 AAF53442.5 AHN54467.1 KMY90346.1 CH933807 EDW11769.1 CH954177 EDV58005.1 KRJ98692.1 KRG02903.1 KQS70221.1 CH963913 EDW77486.2 CVRI01000058 CRL02651.1 JRES01000933 KNC27091.1 CH480818 EDW51808.1 ABLF02036257 KRT03480.1 AAAB01008984 EAA14838.3 KRF81222.1 JXJN01012425 JXJN01012426 JXJN01012427 OUUW01000040 SPP89930.1 UFQS01001542 UFQT01001542 SSX11402.1 SSX30970.1 UFQT01002584 SSX33715.1 CCAG010004606 CH477212 EAT47663.2 KQ434850 KZC08570.1 GL445407 EFN89635.1 DS231846 EDS36168.1 KZ288210 PBC32933.1 ATLV01014217 ATLV01014218 ATLV01014219 ATLV01014220 ATLV01014221 KE524948 KFB38458.1 GGMR01006009 MBY18628.1 KQ435711 KOX79465.1 AXCN02000623 APCN01003424 KK107218 QOIP01000001 EZA55162.1 RLU26481.1 GEDC01024952 JAS12346.1 ADMH02000956 ETN64579.1 ADTU01002189 KQ981042 KYN10101.1 GL439470 EFN67258.1 AXCM01000503 KQ978344 KYM94855.1 GL888424 EGI61143.1 KQ976716 KYM76840.1 AAZX01004127 KQ981805 KYN35630.1 KQ982431 KYQ56500.1 AJWK01035450 AJWK01035451
PZC72541.1 ODYU01006995 SOQ49347.1 AGBW02014188 OWR42121.1 NWSH01001061 PCG72850.1 RSAL01000168 RVE45242.1 KQ459232 KPJ02766.1 DS235252 EEB14009.1 AGBW02012643 OWR44663.1 JTDY01000063 KOB79092.1 KQ971321 EFA01333.2 NEVH01024535 PNF16900.1 PYGN01000441 PSN46305.1 KK853075 KDR11757.1 GFXV01002689 MBW14494.1 GGMS01015681 MBY84884.1 CH916368 EDW02850.1 CH379058 KRT03481.1 CH479184 EDW37056.1 EAL34415.3 KRT03478.1 KRT03479.1 CH940649 EDW63645.1 GECZ01009379 JAS60390.1 KRF81223.1 CH902620 EDV31854.2 CM002910 KMY90345.1 CM000157 EDW89771.2 AJVK01005457 AJVK01005458 AJVK01005459 AJVK01005460 KPU73660.1 KPU73659.1 AE014134 AAF53442.5 AHN54467.1 KMY90346.1 CH933807 EDW11769.1 CH954177 EDV58005.1 KRJ98692.1 KRG02903.1 KQS70221.1 CH963913 EDW77486.2 CVRI01000058 CRL02651.1 JRES01000933 KNC27091.1 CH480818 EDW51808.1 ABLF02036257 KRT03480.1 AAAB01008984 EAA14838.3 KRF81222.1 JXJN01012425 JXJN01012426 JXJN01012427 OUUW01000040 SPP89930.1 UFQS01001542 UFQT01001542 SSX11402.1 SSX30970.1 UFQT01002584 SSX33715.1 CCAG010004606 CH477212 EAT47663.2 KQ434850 KZC08570.1 GL445407 EFN89635.1 DS231846 EDS36168.1 KZ288210 PBC32933.1 ATLV01014217 ATLV01014218 ATLV01014219 ATLV01014220 ATLV01014221 KE524948 KFB38458.1 GGMR01006009 MBY18628.1 KQ435711 KOX79465.1 AXCN02000623 APCN01003424 KK107218 QOIP01000001 EZA55162.1 RLU26481.1 GEDC01024952 JAS12346.1 ADMH02000956 ETN64579.1 ADTU01002189 KQ981042 KYN10101.1 GL439470 EFN67258.1 AXCM01000503 KQ978344 KYM94855.1 GL888424 EGI61143.1 KQ976716 KYM76840.1 AAZX01004127 KQ981805 KYN35630.1 KQ982431 KYQ56500.1 AJWK01035450 AJWK01035451
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000283053
UP000053268
+ More
UP000009046 UP000037510 UP000007266 UP000235965 UP000245037 UP000027135 UP000001070 UP000001819 UP000008744 UP000008792 UP000007801 UP000002282 UP000092462 UP000000803 UP000009192 UP000192221 UP000008711 UP000007798 UP000183832 UP000037069 UP000001292 UP000007819 UP000095301 UP000007062 UP000091820 UP000092443 UP000095300 UP000092460 UP000078200 UP000268350 UP000192223 UP000092444 UP000092445 UP000008820 UP000076502 UP000008237 UP000002320 UP000242457 UP000030765 UP000005203 UP000075885 UP000076408 UP000053105 UP000069272 UP000075886 UP000075840 UP000075902 UP000075900 UP000053097 UP000279307 UP000076407 UP000075884 UP000075882 UP000075880 UP000075920 UP000000673 UP000005205 UP000078492 UP000000311 UP000075883 UP000078542 UP000007755 UP000078540 UP000002358 UP000078541 UP000075809 UP000092461
UP000009046 UP000037510 UP000007266 UP000235965 UP000245037 UP000027135 UP000001070 UP000001819 UP000008744 UP000008792 UP000007801 UP000002282 UP000092462 UP000000803 UP000009192 UP000192221 UP000008711 UP000007798 UP000183832 UP000037069 UP000001292 UP000007819 UP000095301 UP000007062 UP000091820 UP000092443 UP000095300 UP000092460 UP000078200 UP000268350 UP000192223 UP000092444 UP000092445 UP000008820 UP000076502 UP000008237 UP000002320 UP000242457 UP000030765 UP000005203 UP000075885 UP000076408 UP000053105 UP000069272 UP000075886 UP000075840 UP000075902 UP000075900 UP000053097 UP000279307 UP000076407 UP000075884 UP000075882 UP000075880 UP000075920 UP000000673 UP000005205 UP000078492 UP000000311 UP000075883 UP000078542 UP000007755 UP000078540 UP000002358 UP000078541 UP000075809 UP000092461
Pfam
Interpro
IPR032675
LRR_dom_sf
+ More
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001633 EAL_dom
IPR026906 LRR_5
IPR000483 Cys-rich_flank_reg_C
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR005814 Aminotrans_3
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR032135 DUF4817
IPR025875 Leu-rich_rpt_4
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR001633 EAL_dom
IPR026906 LRR_5
IPR000483 Cys-rich_flank_reg_C
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR005814 Aminotrans_3
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR032135 DUF4817
IPR025875 Leu-rich_rpt_4
SUPFAM
SSF53383
SSF53383
Gene 3D
ProteinModelPortal
H9J843
A0A0N0PE55
A0A2A4JKM1
A0A2W1BDV4
A0A2H1W8H3
A0A212EKW8
+ More
A0A2A4JN70 A0A3S2LVH4 A0A194QH46 E0VKV3 A0A212ET41 A0A0L7LUG8 D6WHI0 A0A2J7PKP3 A0A2P8YPX1 A0A067QSJ1 A0A2H8TK33 A0A2S2R4K0 B4JAY0 A0A0R3NR54 B4GK90 Q29P78 B4LRW9 A0A1B6GDA1 A0A0Q9WG37 B3MM94 A0A0J9TNN6 B4NYD5 A0A1B0DDM6 A0A0N8NZ90 A0A0P8XG79 Q9VJQ0 A0A0J9R2M7 B4KLA8 A0A1W4VY78 B3N5P6 A0A0R1DN70 A0A0Q9XFX5 A0A0Q5W9N8 B4MZE7 A0A1J1IQY4 A0A0L0C4D4 B4HXM8 J9JTX0 A0A1I8M578 A0A0R3NQR3 Q7PWD3 A0A1A9WZG1 A0A1A9XY19 A0A0Q9WA29 A0A1I8QBR3 A0A1I8QBR1 A0A1B0BDC5 A0A1A9UH23 A0A3B0KMR9 A0A336MLY2 A0A1W4WPN6 A0A336MVI4 A0A1B0FRG7 A0A1A9Z997 Q17LV0 A0A1S4EYA8 A0A154PBK7 E2B3W6 B0W620 A0A2A3EMA2 A0A084VKG4 A0A088AAQ5 A0A182P6W4 A0A2S2NNB4 A0A182YFT1 A0A0M9A8L2 A0A182F265 A0A182PZJ5 A0A182I7L9 A0A182TZN0 A0A182R6L0 A0A026WGI9 A0A182WXQ5 A0A182NBF6 A0A182LMF4 A0A182IXJ5 A0A182VU11 A0A1S4GZU6 A0A1B6CFS9 W5JP87 A0A158NLR7 A0A195DB55 E2AH27 A0A182LWN1 A0A195C2N7 F4WXH7 A0A195AXS6 K7IWS3 A0A195F4W9 A0A151X857 A0A1B0CY43
A0A2A4JN70 A0A3S2LVH4 A0A194QH46 E0VKV3 A0A212ET41 A0A0L7LUG8 D6WHI0 A0A2J7PKP3 A0A2P8YPX1 A0A067QSJ1 A0A2H8TK33 A0A2S2R4K0 B4JAY0 A0A0R3NR54 B4GK90 Q29P78 B4LRW9 A0A1B6GDA1 A0A0Q9WG37 B3MM94 A0A0J9TNN6 B4NYD5 A0A1B0DDM6 A0A0N8NZ90 A0A0P8XG79 Q9VJQ0 A0A0J9R2M7 B4KLA8 A0A1W4VY78 B3N5P6 A0A0R1DN70 A0A0Q9XFX5 A0A0Q5W9N8 B4MZE7 A0A1J1IQY4 A0A0L0C4D4 B4HXM8 J9JTX0 A0A1I8M578 A0A0R3NQR3 Q7PWD3 A0A1A9WZG1 A0A1A9XY19 A0A0Q9WA29 A0A1I8QBR3 A0A1I8QBR1 A0A1B0BDC5 A0A1A9UH23 A0A3B0KMR9 A0A336MLY2 A0A1W4WPN6 A0A336MVI4 A0A1B0FRG7 A0A1A9Z997 Q17LV0 A0A1S4EYA8 A0A154PBK7 E2B3W6 B0W620 A0A2A3EMA2 A0A084VKG4 A0A088AAQ5 A0A182P6W4 A0A2S2NNB4 A0A182YFT1 A0A0M9A8L2 A0A182F265 A0A182PZJ5 A0A182I7L9 A0A182TZN0 A0A182R6L0 A0A026WGI9 A0A182WXQ5 A0A182NBF6 A0A182LMF4 A0A182IXJ5 A0A182VU11 A0A1S4GZU6 A0A1B6CFS9 W5JP87 A0A158NLR7 A0A195DB55 E2AH27 A0A182LWN1 A0A195C2N7 F4WXH7 A0A195AXS6 K7IWS3 A0A195F4W9 A0A151X857 A0A1B0CY43
PDB
4KT1
E-value=2.18236e-27,
Score=309
Ontologies
GO
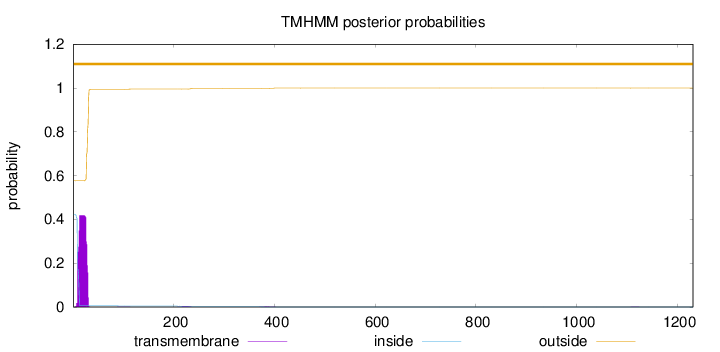
Topology
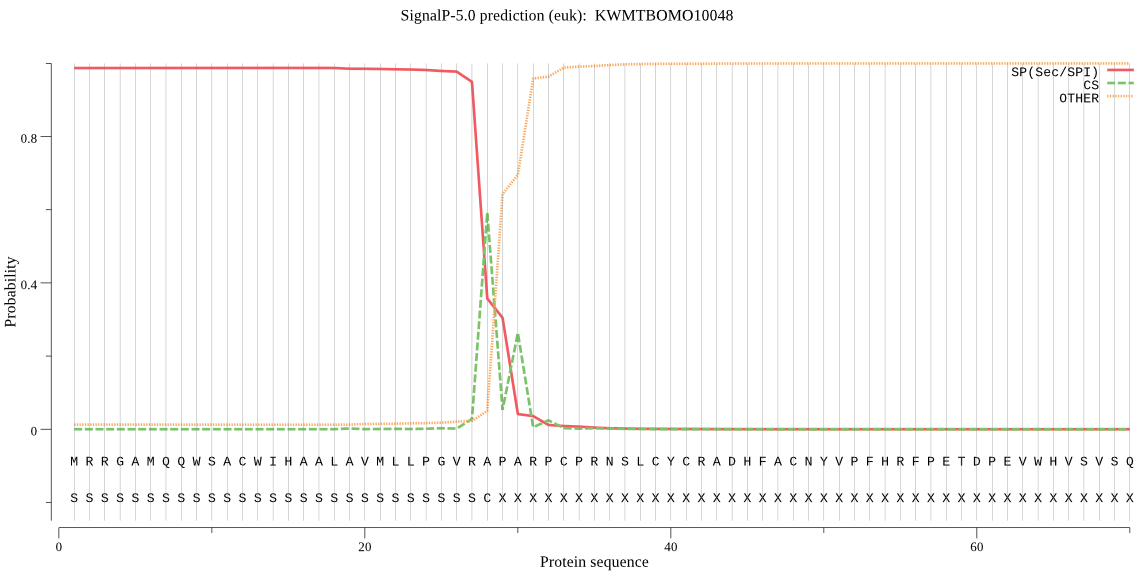
SignalP
Position: 1 - 28,
Likelihood: 0.987354
Length:
1231
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.1354599999999
Exp number, first 60 AAs:
7.99497
Total prob of N-in:
0.42331
outside
1 - 1231
Population Genetic Test Statistics
Pi
187.226744
Theta
160.147664
Tajima's D
0.122241
CLR
0.335894
CSRT
0.408879556022199
Interpretation
Uncertain
