Pre Gene Modal
BGIBMGA005686
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_tuberin_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.745 PlasmaMembrane Reliability : 1.139
Sequence
CDS
ATGGGGTCTAGGGACAGGGACTCGAAATCGCTACAGGACAAGCTGAAAGTGTTCTTCAAGAAAGGTGCATCGGCGCCGCTAGCGGCTCGCAGCGAGCTGGTGTCGAAGGAGGTGCTTCATGAGTTGAGCGTCGAAACGCCGCTGCACCGACGGCTCCGTGCCATGAAGGATGTTGGCGACAAGGTCATTCAGGGACGCGTGGAAGACGGTGGTGTTGAAAAGTTATGGTCTTGCACTCGAGACCTCCTCTACACGGATAATACCGAAGCCCGTCACACAGCCCTGTGTTTCATGAGATGTATCGCTGAAGGACAAGCAGAAGATCTACAAATAATGAGGACAATTTTCTTCAAATTCCTAAGGGAGACACACCCGTCTCATCCGCCAGACGACTATCAATTACGCTTTAAACTACTCTATACATTGACTAATAGCGGGAAAAATATTAATTGTTTTGAAGTGGATATTGGAGAGTTCTTATTGGAATGGCTGCCTCAAATTCAGACACCATCAAATACAGTTGAATTTTTACAGCTAATCGTAAATGTAGTAAAATTCAATGCTACATATCTGGACGAACACATTGTACATGGAATTGTCAACAACGCATGCCACTTGTGCGTATATTCCCCGGAGGTGACAGTTGTGCAGCAGTGCCTCTCCTTGCTCGAGGCTGTCGTGTCGTACTCGCTGTTGCCCCGGGCGTCCCTGCCGACGTTTGTCGGTGCGCTGTGCCGGACTGTCAACATGGAACACTACTGTCAAAATAGTTGGAAGTTGATGCGGTACGTGGTGGGCGGAGACATCGGTCCGGCGGCCGTGCAGCAGATGGTGTCGCTGCTGCAGTCGGGGGCGGAGGCCGGGGCTGCCAGGGGCGCCGTGTTCTATATCAACATGGCACTGTGGGGGCCGCAACGTGTGCCCACGCTCAAGGTGTCATTTCTGGCCGTCCTACCGGCCTTTCTGAAGGCGCTGGAGGGTCGCCAGGCGGTGGTGGCGTACGAGGTGGTGGTGGGGGTGGCCAGCGTGGTCACGCGAGCCCCCCACGAGCTGTACGAGCCGGCCTGGGACGTGCTGCTGAGGATCGTGCGAACCCTGCTGCAGCTCGACAAATCGTACGACCCCCCCAACGACTTGATGCACTCCCGGCTGCACAACATCATAACGTCGATCGAACAACTCCAGGAGTCCGGCCAGTACAAGGGCGACGCGGAGGCCCTGCTGGACCTCGTGGACAGCTGCGGACACGAGCGCCCGTTGACCCTTCAGACTTCGTAG
Protein
MGSRDRDSKSLQDKLKVFFKKGASAPLAARSELVSKEVLHELSVETPLHRRLRAMKDVGDKVIQGRVEDGGVEKLWSCTRDLLYTDNTEARHTALCFMRCIAEGQAEDLQIMRTIFFKFLRETHPSHPPDDYQLRFKLLYTLTNSGKNINCFEVDIGEFLLEWLPQIQTPSNTVEFLQLIVNVVKFNATYLDEHIVHGIVNNACHLCVYSPEVTVVQQCLSLLEAVVSYSLLPRASLPTFVGALCRTVNMEHYCQNSWKLMRYVVGGDIGPAAVQQMVSLLQSGAEAGAARGAVFYINMALWGPQRVPTLKVSFLAVLPAFLKALEGRQAVVAYEVVVGVASVVTRAPHELYEPAWDVLLRIVRTLLQLDKSYDPPNDLMHSRLHNIITSIEQLQESGQYKGDAEALLDLVDSCGHERPLTLQTS
Summary
Uniprot
A0A2W1BH62
A0A2A4JAJ7
A0A212FF76
A0A194QAG3
A0A0L7LRT4
A0A0N1IQ05
+ More
A0A2H1VXQ9 A0A2P8YFZ1 A0A2J7PKX2 A0A067R1A4 A0A1B6CHU9 A0A0N7Z993 A0A0C9QWE4 A0A154P5U1 A0A2A3E2S5 A0A088ADA5 A0A026VUB2 A0A151WRN0 A0A158NUP5 F4WRA9 A0A151IG77 A0A0M4M1C4 A0A151IS99 A0A2B4S1N1 E2AD21 K7IN19 A0A232F0T6 N6TJ61 A0A2R7WND5 A0A1B6KY98 A0A0A9XDX4 A0A1S3HHV4 A0A0K8TBU2 A0A195FK85 C3Z790 U4UEW8 E0VSQ1 A0A195BBK3 A0A3L8DJ68 A0A1B6G2M3 A0A2C9KD24 A0A2R8YEJ8 A0A1V1FNX1 X5DRC8 K1QPU2 D6WZ21 A0A3B3REN6 A0A3B3RCX5 A0A3B3RD40 A0A3B3RCF9 A0A287A4I9 F1RFB2 A0A287ACI2 A0A287AG89 A0A287ADS4 A0A287BTB9 A0A2K6RI22 X5D7Z6 A0A287BBA5 A0A287BG63 X5D9K3 A0A2R9B2X2 Q3UI96 A0A1V1FFQ4 A0A2K5ZR95 A0A2K5V7X6 A0A3Q2IBY1 A0A1D5QQI8 A0A3Q2H0W3 A0A2I3M4Y1 F6WI30 A0A3Q2L218 A0A091DWS9 A0A2K5MV13 A0A1V1G0D9 F6ZQU1 W5M5E7 W5M5G2 A0A2Y9SP68 A0A2Y9EDW4 A0A1V1FNX9 A0A1V1FI06 A0A2Y9EE39 A0A2K6RI55 A0A2Y9PYI6 A0A2Y9Q480 A0A2Y9PTM1 L5LUJ4 A0A341CM49 A0A2Y9Q484 A0A2K6RI03 A0A2Y9PYI2 A0A341CLG7 A0A341CLE4 A0A2Y9PXK2 A0A1S3WIC2 A0A2K5MVI9 A0A1S3WHM3 A0A1S3A201
A0A2H1VXQ9 A0A2P8YFZ1 A0A2J7PKX2 A0A067R1A4 A0A1B6CHU9 A0A0N7Z993 A0A0C9QWE4 A0A154P5U1 A0A2A3E2S5 A0A088ADA5 A0A026VUB2 A0A151WRN0 A0A158NUP5 F4WRA9 A0A151IG77 A0A0M4M1C4 A0A151IS99 A0A2B4S1N1 E2AD21 K7IN19 A0A232F0T6 N6TJ61 A0A2R7WND5 A0A1B6KY98 A0A0A9XDX4 A0A1S3HHV4 A0A0K8TBU2 A0A195FK85 C3Z790 U4UEW8 E0VSQ1 A0A195BBK3 A0A3L8DJ68 A0A1B6G2M3 A0A2C9KD24 A0A2R8YEJ8 A0A1V1FNX1 X5DRC8 K1QPU2 D6WZ21 A0A3B3REN6 A0A3B3RCX5 A0A3B3RD40 A0A3B3RCF9 A0A287A4I9 F1RFB2 A0A287ACI2 A0A287AG89 A0A287ADS4 A0A287BTB9 A0A2K6RI22 X5D7Z6 A0A287BBA5 A0A287BG63 X5D9K3 A0A2R9B2X2 Q3UI96 A0A1V1FFQ4 A0A2K5ZR95 A0A2K5V7X6 A0A3Q2IBY1 A0A1D5QQI8 A0A3Q2H0W3 A0A2I3M4Y1 F6WI30 A0A3Q2L218 A0A091DWS9 A0A2K5MV13 A0A1V1G0D9 F6ZQU1 W5M5E7 W5M5G2 A0A2Y9SP68 A0A2Y9EDW4 A0A1V1FNX9 A0A1V1FI06 A0A2Y9EE39 A0A2K6RI55 A0A2Y9PYI6 A0A2Y9Q480 A0A2Y9PTM1 L5LUJ4 A0A341CM49 A0A2Y9Q484 A0A2K6RI03 A0A2Y9PYI2 A0A341CLG7 A0A341CLE4 A0A2Y9PXK2 A0A1S3WIC2 A0A2K5MVI9 A0A1S3WHM3 A0A1S3A201
Pubmed
28756777
22118469
26354079
26227816
29403074
24845553
+ More
27129103 24508170 21347285 21719571 20798317 20075255 28648823 23537049 25401762 18563158 20566863 30249741 15562597 15616553 21269460 24722188 22992520 18362917 19820115 29240929 30723633 25362486 22722832 10349636 11042159 11076861 11217851 12466851 16141073 19892987 17431167 18464734
27129103 24508170 21347285 21719571 20798317 20075255 28648823 23537049 25401762 18563158 20566863 30249741 15562597 15616553 21269460 24722188 22992520 18362917 19820115 29240929 30723633 25362486 22722832 10349636 11042159 11076861 11217851 12466851 16141073 19892987 17431167 18464734
EMBL
KZ150179
PZC72537.1
NWSH01002371
PCG68450.1
AGBW02008849
OWR52395.1
+ More
KQ459232 KPJ02523.1 JTDY01000229 KOB78178.1 KQ460044 KPJ18291.1 ODYU01005093 SOQ45627.1 PYGN01000627 PSN43169.1 NEVH01024531 PNF16986.1 KK852784 KDR16585.1 GEDC01024280 JAS13018.1 GDKW01001187 JAI55408.1 GBYB01000035 JAG69802.1 KQ434823 KZC07296.1 KZ288467 PBC25401.1 KK107942 EZA47091.1 KQ982806 KYQ50457.1 ADTU01026650 ADTU01026651 ADTU01026652 GL888284 EGI63326.1 KQ977739 KYN00227.1 KR075843 ALE20562.1 KQ981085 KYN09595.1 LSMT01000223 PFX22959.1 GL438658 EFN68657.1 NNAY01001310 OXU24396.1 APGK01035639 KB740928 ENN77893.1 KK855147 PTY21138.1 GEBQ01023743 JAT16234.1 GBHO01025435 JAG18169.1 GBRD01003191 JAG62630.1 KQ981512 KYN40813.1 GG666590 EEN51686.1 KB632064 ERL88460.1 DS235755 EEB16407.1 KQ976532 KYM81590.1 QOIP01000007 RLU20252.1 GECZ01013082 JAS56687.1 AC093513 LC224282 BAX02962.1 KJ534982 AHW56622.1 JH818015 EKC33134.1 KQ971363 EFA09042.2 AEMK02000018 DQIR01165177 HDB20654.1 KJ535091 AHW56730.1 KJ535039 AHW56678.1 AJFE02022693 AJFE02022694 AK147016 BAE27610.1 LC224288 BAX02968.1 AQIA01041640 AQIA01041641 AQIA01041642 AQIA01041643 AQIA01041644 JSUE03025444 JSUE03025445 JSUE03025446 JSUE03025447 AHZZ02018195 KN121905 KFO34715.1 LC224283 LC224290 BAX02963.1 AHAT01037171 LC224292 BAX02972.1 LC224289 BAX02969.1 KB107323 ELK30094.1
KQ459232 KPJ02523.1 JTDY01000229 KOB78178.1 KQ460044 KPJ18291.1 ODYU01005093 SOQ45627.1 PYGN01000627 PSN43169.1 NEVH01024531 PNF16986.1 KK852784 KDR16585.1 GEDC01024280 JAS13018.1 GDKW01001187 JAI55408.1 GBYB01000035 JAG69802.1 KQ434823 KZC07296.1 KZ288467 PBC25401.1 KK107942 EZA47091.1 KQ982806 KYQ50457.1 ADTU01026650 ADTU01026651 ADTU01026652 GL888284 EGI63326.1 KQ977739 KYN00227.1 KR075843 ALE20562.1 KQ981085 KYN09595.1 LSMT01000223 PFX22959.1 GL438658 EFN68657.1 NNAY01001310 OXU24396.1 APGK01035639 KB740928 ENN77893.1 KK855147 PTY21138.1 GEBQ01023743 JAT16234.1 GBHO01025435 JAG18169.1 GBRD01003191 JAG62630.1 KQ981512 KYN40813.1 GG666590 EEN51686.1 KB632064 ERL88460.1 DS235755 EEB16407.1 KQ976532 KYM81590.1 QOIP01000007 RLU20252.1 GECZ01013082 JAS56687.1 AC093513 LC224282 BAX02962.1 KJ534982 AHW56622.1 JH818015 EKC33134.1 KQ971363 EFA09042.2 AEMK02000018 DQIR01165177 HDB20654.1 KJ535091 AHW56730.1 KJ535039 AHW56678.1 AJFE02022693 AJFE02022694 AK147016 BAE27610.1 LC224288 BAX02968.1 AQIA01041640 AQIA01041641 AQIA01041642 AQIA01041643 AQIA01041644 JSUE03025444 JSUE03025445 JSUE03025446 JSUE03025447 AHZZ02018195 KN121905 KFO34715.1 LC224283 LC224290 BAX02963.1 AHAT01037171 LC224292 BAX02972.1 LC224289 BAX02969.1 KB107323 ELK30094.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000037510
UP000053240
UP000245037
+ More
UP000235965 UP000027135 UP000076502 UP000242457 UP000005203 UP000053097 UP000075809 UP000005205 UP000007755 UP000078542 UP000078492 UP000225706 UP000000311 UP000002358 UP000215335 UP000019118 UP000085678 UP000078541 UP000001554 UP000030742 UP000009046 UP000078540 UP000279307 UP000076420 UP000005640 UP000005408 UP000007266 UP000261540 UP000008227 UP000233200 UP000240080 UP000233140 UP000233100 UP000002281 UP000006718 UP000028761 UP000028990 UP000233060 UP000002279 UP000018468 UP000248484 UP000248483 UP000252040 UP000079721
UP000235965 UP000027135 UP000076502 UP000242457 UP000005203 UP000053097 UP000075809 UP000005205 UP000007755 UP000078542 UP000078492 UP000225706 UP000000311 UP000002358 UP000215335 UP000019118 UP000085678 UP000078541 UP000001554 UP000030742 UP000009046 UP000078540 UP000279307 UP000076420 UP000005640 UP000005408 UP000007266 UP000261540 UP000008227 UP000233200 UP000240080 UP000233140 UP000233100 UP000002281 UP000006718 UP000028761 UP000028990 UP000233060 UP000002279 UP000018468 UP000248484 UP000248483 UP000252040 UP000079721
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BH62
A0A2A4JAJ7
A0A212FF76
A0A194QAG3
A0A0L7LRT4
A0A0N1IQ05
+ More
A0A2H1VXQ9 A0A2P8YFZ1 A0A2J7PKX2 A0A067R1A4 A0A1B6CHU9 A0A0N7Z993 A0A0C9QWE4 A0A154P5U1 A0A2A3E2S5 A0A088ADA5 A0A026VUB2 A0A151WRN0 A0A158NUP5 F4WRA9 A0A151IG77 A0A0M4M1C4 A0A151IS99 A0A2B4S1N1 E2AD21 K7IN19 A0A232F0T6 N6TJ61 A0A2R7WND5 A0A1B6KY98 A0A0A9XDX4 A0A1S3HHV4 A0A0K8TBU2 A0A195FK85 C3Z790 U4UEW8 E0VSQ1 A0A195BBK3 A0A3L8DJ68 A0A1B6G2M3 A0A2C9KD24 A0A2R8YEJ8 A0A1V1FNX1 X5DRC8 K1QPU2 D6WZ21 A0A3B3REN6 A0A3B3RCX5 A0A3B3RD40 A0A3B3RCF9 A0A287A4I9 F1RFB2 A0A287ACI2 A0A287AG89 A0A287ADS4 A0A287BTB9 A0A2K6RI22 X5D7Z6 A0A287BBA5 A0A287BG63 X5D9K3 A0A2R9B2X2 Q3UI96 A0A1V1FFQ4 A0A2K5ZR95 A0A2K5V7X6 A0A3Q2IBY1 A0A1D5QQI8 A0A3Q2H0W3 A0A2I3M4Y1 F6WI30 A0A3Q2L218 A0A091DWS9 A0A2K5MV13 A0A1V1G0D9 F6ZQU1 W5M5E7 W5M5G2 A0A2Y9SP68 A0A2Y9EDW4 A0A1V1FNX9 A0A1V1FI06 A0A2Y9EE39 A0A2K6RI55 A0A2Y9PYI6 A0A2Y9Q480 A0A2Y9PTM1 L5LUJ4 A0A341CM49 A0A2Y9Q484 A0A2K6RI03 A0A2Y9PYI2 A0A341CLG7 A0A341CLE4 A0A2Y9PXK2 A0A1S3WIC2 A0A2K5MVI9 A0A1S3WHM3 A0A1S3A201
A0A2H1VXQ9 A0A2P8YFZ1 A0A2J7PKX2 A0A067R1A4 A0A1B6CHU9 A0A0N7Z993 A0A0C9QWE4 A0A154P5U1 A0A2A3E2S5 A0A088ADA5 A0A026VUB2 A0A151WRN0 A0A158NUP5 F4WRA9 A0A151IG77 A0A0M4M1C4 A0A151IS99 A0A2B4S1N1 E2AD21 K7IN19 A0A232F0T6 N6TJ61 A0A2R7WND5 A0A1B6KY98 A0A0A9XDX4 A0A1S3HHV4 A0A0K8TBU2 A0A195FK85 C3Z790 U4UEW8 E0VSQ1 A0A195BBK3 A0A3L8DJ68 A0A1B6G2M3 A0A2C9KD24 A0A2R8YEJ8 A0A1V1FNX1 X5DRC8 K1QPU2 D6WZ21 A0A3B3REN6 A0A3B3RCX5 A0A3B3RD40 A0A3B3RCF9 A0A287A4I9 F1RFB2 A0A287ACI2 A0A287AG89 A0A287ADS4 A0A287BTB9 A0A2K6RI22 X5D7Z6 A0A287BBA5 A0A287BG63 X5D9K3 A0A2R9B2X2 Q3UI96 A0A1V1FFQ4 A0A2K5ZR95 A0A2K5V7X6 A0A3Q2IBY1 A0A1D5QQI8 A0A3Q2H0W3 A0A2I3M4Y1 F6WI30 A0A3Q2L218 A0A091DWS9 A0A2K5MV13 A0A1V1G0D9 F6ZQU1 W5M5E7 W5M5G2 A0A2Y9SP68 A0A2Y9EDW4 A0A1V1FNX9 A0A1V1FI06 A0A2Y9EE39 A0A2K6RI55 A0A2Y9PYI6 A0A2Y9Q480 A0A2Y9PTM1 L5LUJ4 A0A341CM49 A0A2Y9Q484 A0A2K6RI03 A0A2Y9PYI2 A0A341CLG7 A0A341CLE4 A0A2Y9PXK2 A0A1S3WIC2 A0A2K5MVI9 A0A1S3WHM3 A0A1S3A201
PDB
5HIU
E-value=1.1892e-06,
Score=126
Ontologies
PATHWAY
GO
GO:0033596
GO:0051056
GO:0005096
GO:0032007
GO:0005634
GO:0016021
GO:0046627
GO:0051726
GO:0030178
GO:0005737
GO:0051898
GO:0048471
GO:0042803
GO:0016020
GO:0031267
GO:0014069
GO:0043276
GO:0005764
GO:0051879
GO:0005794
GO:0016239
GO:0019902
GO:0005488
GO:0043547
GO:0004672
GO:0030677
GO:0016155
GO:0003824
PANTHER
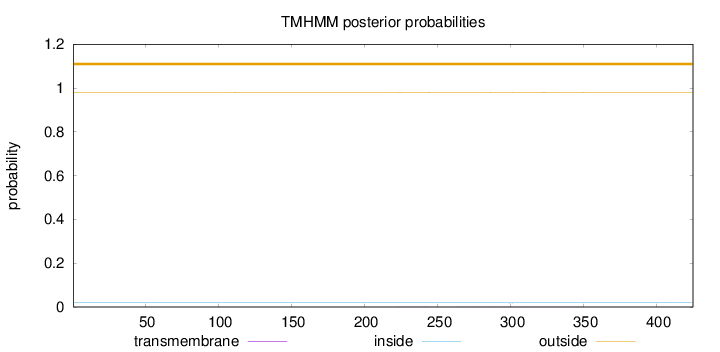
Topology
Length:
425
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04245
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01957
outside
1 - 425
Population Genetic Test Statistics
Pi
141.112774
Theta
140.856418
Tajima's D
-0.760019
CLR
0.15025
CSRT
0.186490675466227
Interpretation
Uncertain
