Pre Gene Modal
BGIBMGA005530
Annotation
PREDICTED:_28S_ribosomal_protein_S33?_mitochondrial_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.384
Sequence
CDS
ATGGCCACAAATTATACAAAATATGCTCAACTAATTAATACGTCGACAACGTACGCGCGACGAATGAAGCGCCTGTCCAATAGGATCTTTGGCGAAGTTGCCCTACCTACCAACTCCAAGTCCATGAAGGTTGTCAAAATCTTCTCTGCTCGACCTCTGCACACAAATGAGGAAATTATACACTATTATCCTAGACACATGGAGACGCATAAACTGATGATGAATTTAAGAGATTATGGTCTATTCCGTGATGAGCATCAAGACTTCAAGGAGGAGATGAAGAGATTGCGAGAACTCCGAGGTAAAGTCAAGGTTTGGAGGCGGTTGCTTGATAAGAAGGCGGAAGCAAAAGAATAA
Protein
MATNYTKYAQLINTSTTYARRMKRLSNRIFGEVALPTNSKSMKVVKIFSARPLHTNEEIIHYYPRHMETHKLMMNLRDYGLFRDEHQDFKEEMKRLRELRGKVKVWRRLLDKKAEAKE
Summary
Uniprot
Q2F5Y5
A0A2W1BFL1
A0A2A4JI59
H9J7N8
A0A0L7LRT8
A0A212FF95
+ More
A0A1E1WKR0 S4PML9 A0A194QAS0 A0A0N0PDV2 U5ETH0 A0A0K8TS94 A0A1Y1KRT1 A0A2P8YFU0 A0A2M4AWS2 D6WX59 A0A067R0Z5 A0A2J7R3E6 A0A182RPE3 W5JWY4 A0A2M3ZDD6 A0A2M4AWU1 A0A182F719 A0A2M4C480 A0A182ULP0 A0A182JZ11 A0A182UGR0 A0A182XIS7 A0A182LBW9 Q7PR61 A0A182HTI9 T1DPZ6 Q9VEV5 A0A1Q3G4A9 A0A182VRU3 A0A3B0JQT7 Q295W3 B4GFL6 A0A084W348 A0A182QK22 A0A182LWT1 B4QX34 B4PQZ7 B4IBW0 B3P122 A0A182J0Z1 A0A1W4U7T6 B4KBZ6 A0A195C1U7 B0XG83 A0A023ED59 A0A182N7B7 B4LX41 A0A195AXS1 A0A182PE03 B3M2J7 A0A1I8NI56 Q0IFW2 B4NLA1 A0A195DB01 A0A1L8DMK6 A0A1I8PKG2 A0A0M4ERD4 K7IN17 J9JNG9 A0A158NLU9 A0A1L8EDI6 N6UGX2 A0A232F128 A0A1W4WRC6 A0A151X7Y8 A0A1B0CEE5 A0A195F4X4 B4JFL2 B5BV16 A0A034WWK1 A0A0C9QHW2 A0A0K8V4A6 A0A2H8TIU1 A0A0N0BJQ5 A0A0J7KYB7 E2AF96 A0A1A9WFN9 E9JBY1 A0A1B6DAT2 A0A0P6B4U6 A0A0P5Z7C1 A0A0A1X386 W8BZ50 A0A1B0G4T4 A0A1A9VD31 A0A0P5V7G9 A0A0L0BQI8 A0A1A9ZAD1 A0A0P5DZX8 A0A0P5T8U7 A0A1A9XBD3 A0A0P4WKQ5 A0A154PMU1
A0A1E1WKR0 S4PML9 A0A194QAS0 A0A0N0PDV2 U5ETH0 A0A0K8TS94 A0A1Y1KRT1 A0A2P8YFU0 A0A2M4AWS2 D6WX59 A0A067R0Z5 A0A2J7R3E6 A0A182RPE3 W5JWY4 A0A2M3ZDD6 A0A2M4AWU1 A0A182F719 A0A2M4C480 A0A182ULP0 A0A182JZ11 A0A182UGR0 A0A182XIS7 A0A182LBW9 Q7PR61 A0A182HTI9 T1DPZ6 Q9VEV5 A0A1Q3G4A9 A0A182VRU3 A0A3B0JQT7 Q295W3 B4GFL6 A0A084W348 A0A182QK22 A0A182LWT1 B4QX34 B4PQZ7 B4IBW0 B3P122 A0A182J0Z1 A0A1W4U7T6 B4KBZ6 A0A195C1U7 B0XG83 A0A023ED59 A0A182N7B7 B4LX41 A0A195AXS1 A0A182PE03 B3M2J7 A0A1I8NI56 Q0IFW2 B4NLA1 A0A195DB01 A0A1L8DMK6 A0A1I8PKG2 A0A0M4ERD4 K7IN17 J9JNG9 A0A158NLU9 A0A1L8EDI6 N6UGX2 A0A232F128 A0A1W4WRC6 A0A151X7Y8 A0A1B0CEE5 A0A195F4X4 B4JFL2 B5BV16 A0A034WWK1 A0A0C9QHW2 A0A0K8V4A6 A0A2H8TIU1 A0A0N0BJQ5 A0A0J7KYB7 E2AF96 A0A1A9WFN9 E9JBY1 A0A1B6DAT2 A0A0P6B4U6 A0A0P5Z7C1 A0A0A1X386 W8BZ50 A0A1B0G4T4 A0A1A9VD31 A0A0P5V7G9 A0A0L0BQI8 A0A1A9ZAD1 A0A0P5DZX8 A0A0P5T8U7 A0A1A9XBD3 A0A0P4WKQ5 A0A154PMU1
Pubmed
28756777
19121390
26227816
22118469
23622113
26354079
+ More
26369729 28004739 29403074 18362917 19820115 24845553 20920257 23761445 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17994087 24438588 17550304 24945155 25315136 17510324 20075255 21347285 23537049 28648823 25348373 20798317 21282665 25830018 24495485 26108605
26369729 28004739 29403074 18362917 19820115 24845553 20920257 23761445 20966253 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17994087 24438588 17550304 24945155 25315136 17510324 20075255 21347285 23537049 28648823 25348373 20798317 21282665 25830018 24495485 26108605
EMBL
DQ311287
ABD36232.2
KZ150179
PZC72535.1
NWSH01001312
PCG71747.1
+ More
BABH01020426 JTDY01000229 KOB78183.1 AGBW02008849 OWR52393.1 GDQN01009938 GDQN01003472 JAT81116.1 JAT87582.1 GAIX01000106 JAA92454.1 KQ459232 KPJ02529.1 KQ460044 KPJ18295.1 GANO01002783 JAB57088.1 GDAI01000587 JAI17016.1 GEZM01077465 JAV63298.1 PYGN01000629 PSN43126.1 GGFK01011925 MBW45246.1 KQ971361 EFA08795.1 KK852784 KDR16579.1 NEVH01007821 PNF35340.1 ADMH02000081 ETN67870.1 GGFM01005803 MBW26554.1 GGFK01011945 MBW45266.1 GGFJ01010928 MBW60069.1 AAAB01008859 EAA07794.3 APCN01000536 GAMD01002894 JAA98696.1 AE014297 BT016004 KX531437 AAF55312.1 AAV36889.1 ANY27247.1 GFDL01000400 JAV34645.1 OUUW01000008 SPP84537.1 CM000070 EAL28595.1 CH479182 EDW34401.1 ATLV01019815 KE525279 KFB44642.1 AXCN02000146 AXCM01004364 CM000364 EDX12688.1 CM000160 EDW97319.1 CH480827 EDW44868.1 CH954181 EDV49141.1 CH933806 EDW15848.1 KQ978344 KYM94849.1 DS232999 EDS27301.1 GAPW01006267 GAPW01006266 JAC07332.1 CH940650 EDW66693.1 KQ976716 KYM76835.1 CH902617 EDV43450.1 CH477286 EAT44738.1 CH964272 EDW84304.1 KQ981042 KYN10095.1 GFDF01006490 JAV07594.1 CP012526 ALC47714.1 ABLF02014729 ABLF02014730 ABLF02014731 ADTU01002193 GFDG01002104 JAV16695.1 APGK01035639 KB740928 KB632064 ENN77897.1 ERL88458.1 NNAY01001310 OXU24401.1 KQ982431 KYQ56495.1 AJWK01008791 KQ981805 KYN35635.1 CH916369 EDV93493.1 AB456740 BAG70406.1 GAKP01000352 JAC58600.1 GBYB01014253 JAG84020.1 GDHF01021896 GDHF01018582 JAI30418.1 JAI33732.1 GFXV01001807 GFXV01006132 MBW13612.1 MBW17937.1 KQ435711 KOX79468.1 LBMM01002088 KMQ95284.1 GL441302 GL439067 EFN64932.1 EFN67900.1 GL771642 EFZ09651.1 GEDC01014568 JAS22730.1 GDIP01022093 JAM81622.1 GDIQ01095958 GDIP01047627 LRGB01003333 JAL55768.1 JAM56088.1 KZS03161.1 GBXI01009144 JAD05148.1 GAMC01001974 GAMC01001971 GAMC01001968 JAC04588.1 CCAG010006284 GDIP01103162 JAM00553.1 JRES01001621 KNC21479.1 GDIP01154661 JAJ68741.1 GDIQ01241618 GDIP01129410 GDIP01095262 JAK10107.1 JAL74304.1 GDRN01022700 JAI67779.1 KQ434993 KZC13201.1
BABH01020426 JTDY01000229 KOB78183.1 AGBW02008849 OWR52393.1 GDQN01009938 GDQN01003472 JAT81116.1 JAT87582.1 GAIX01000106 JAA92454.1 KQ459232 KPJ02529.1 KQ460044 KPJ18295.1 GANO01002783 JAB57088.1 GDAI01000587 JAI17016.1 GEZM01077465 JAV63298.1 PYGN01000629 PSN43126.1 GGFK01011925 MBW45246.1 KQ971361 EFA08795.1 KK852784 KDR16579.1 NEVH01007821 PNF35340.1 ADMH02000081 ETN67870.1 GGFM01005803 MBW26554.1 GGFK01011945 MBW45266.1 GGFJ01010928 MBW60069.1 AAAB01008859 EAA07794.3 APCN01000536 GAMD01002894 JAA98696.1 AE014297 BT016004 KX531437 AAF55312.1 AAV36889.1 ANY27247.1 GFDL01000400 JAV34645.1 OUUW01000008 SPP84537.1 CM000070 EAL28595.1 CH479182 EDW34401.1 ATLV01019815 KE525279 KFB44642.1 AXCN02000146 AXCM01004364 CM000364 EDX12688.1 CM000160 EDW97319.1 CH480827 EDW44868.1 CH954181 EDV49141.1 CH933806 EDW15848.1 KQ978344 KYM94849.1 DS232999 EDS27301.1 GAPW01006267 GAPW01006266 JAC07332.1 CH940650 EDW66693.1 KQ976716 KYM76835.1 CH902617 EDV43450.1 CH477286 EAT44738.1 CH964272 EDW84304.1 KQ981042 KYN10095.1 GFDF01006490 JAV07594.1 CP012526 ALC47714.1 ABLF02014729 ABLF02014730 ABLF02014731 ADTU01002193 GFDG01002104 JAV16695.1 APGK01035639 KB740928 KB632064 ENN77897.1 ERL88458.1 NNAY01001310 OXU24401.1 KQ982431 KYQ56495.1 AJWK01008791 KQ981805 KYN35635.1 CH916369 EDV93493.1 AB456740 BAG70406.1 GAKP01000352 JAC58600.1 GBYB01014253 JAG84020.1 GDHF01021896 GDHF01018582 JAI30418.1 JAI33732.1 GFXV01001807 GFXV01006132 MBW13612.1 MBW17937.1 KQ435711 KOX79468.1 LBMM01002088 KMQ95284.1 GL441302 GL439067 EFN64932.1 EFN67900.1 GL771642 EFZ09651.1 GEDC01014568 JAS22730.1 GDIP01022093 JAM81622.1 GDIQ01095958 GDIP01047627 LRGB01003333 JAL55768.1 JAM56088.1 KZS03161.1 GBXI01009144 JAD05148.1 GAMC01001974 GAMC01001971 GAMC01001968 JAC04588.1 CCAG010006284 GDIP01103162 JAM00553.1 JRES01001621 KNC21479.1 GDIP01154661 JAJ68741.1 GDIQ01241618 GDIP01129410 GDIP01095262 JAK10107.1 JAL74304.1 GDRN01022700 JAI67779.1 KQ434993 KZC13201.1
Proteomes
UP000218220
UP000005204
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000245037 UP000007266 UP000027135 UP000235965 UP000075900 UP000000673 UP000069272 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000000803 UP000075920 UP000268350 UP000001819 UP000008744 UP000030765 UP000075886 UP000075883 UP000000304 UP000002282 UP000001292 UP000008711 UP000075880 UP000192221 UP000009192 UP000078542 UP000002320 UP000075884 UP000008792 UP000078540 UP000075885 UP000007801 UP000095301 UP000008820 UP000007798 UP000078492 UP000095300 UP000092553 UP000002358 UP000007819 UP000005205 UP000019118 UP000030742 UP000215335 UP000192223 UP000075809 UP000092461 UP000078541 UP000001070 UP000053105 UP000036403 UP000000311 UP000091820 UP000076858 UP000092444 UP000078200 UP000037069 UP000092445 UP000092443 UP000076502
UP000245037 UP000007266 UP000027135 UP000235965 UP000075900 UP000000673 UP000069272 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000000803 UP000075920 UP000268350 UP000001819 UP000008744 UP000030765 UP000075886 UP000075883 UP000000304 UP000002282 UP000001292 UP000008711 UP000075880 UP000192221 UP000009192 UP000078542 UP000002320 UP000075884 UP000008792 UP000078540 UP000075885 UP000007801 UP000095301 UP000008820 UP000007798 UP000078492 UP000095300 UP000092553 UP000002358 UP000007819 UP000005205 UP000019118 UP000030742 UP000215335 UP000192223 UP000075809 UP000092461 UP000078541 UP000001070 UP000053105 UP000036403 UP000000311 UP000091820 UP000076858 UP000092444 UP000078200 UP000037069 UP000092445 UP000092443 UP000076502
Interpro
ProteinModelPortal
Q2F5Y5
A0A2W1BFL1
A0A2A4JI59
H9J7N8
A0A0L7LRT8
A0A212FF95
+ More
A0A1E1WKR0 S4PML9 A0A194QAS0 A0A0N0PDV2 U5ETH0 A0A0K8TS94 A0A1Y1KRT1 A0A2P8YFU0 A0A2M4AWS2 D6WX59 A0A067R0Z5 A0A2J7R3E6 A0A182RPE3 W5JWY4 A0A2M3ZDD6 A0A2M4AWU1 A0A182F719 A0A2M4C480 A0A182ULP0 A0A182JZ11 A0A182UGR0 A0A182XIS7 A0A182LBW9 Q7PR61 A0A182HTI9 T1DPZ6 Q9VEV5 A0A1Q3G4A9 A0A182VRU3 A0A3B0JQT7 Q295W3 B4GFL6 A0A084W348 A0A182QK22 A0A182LWT1 B4QX34 B4PQZ7 B4IBW0 B3P122 A0A182J0Z1 A0A1W4U7T6 B4KBZ6 A0A195C1U7 B0XG83 A0A023ED59 A0A182N7B7 B4LX41 A0A195AXS1 A0A182PE03 B3M2J7 A0A1I8NI56 Q0IFW2 B4NLA1 A0A195DB01 A0A1L8DMK6 A0A1I8PKG2 A0A0M4ERD4 K7IN17 J9JNG9 A0A158NLU9 A0A1L8EDI6 N6UGX2 A0A232F128 A0A1W4WRC6 A0A151X7Y8 A0A1B0CEE5 A0A195F4X4 B4JFL2 B5BV16 A0A034WWK1 A0A0C9QHW2 A0A0K8V4A6 A0A2H8TIU1 A0A0N0BJQ5 A0A0J7KYB7 E2AF96 A0A1A9WFN9 E9JBY1 A0A1B6DAT2 A0A0P6B4U6 A0A0P5Z7C1 A0A0A1X386 W8BZ50 A0A1B0G4T4 A0A1A9VD31 A0A0P5V7G9 A0A0L0BQI8 A0A1A9ZAD1 A0A0P5DZX8 A0A0P5T8U7 A0A1A9XBD3 A0A0P4WKQ5 A0A154PMU1
A0A1E1WKR0 S4PML9 A0A194QAS0 A0A0N0PDV2 U5ETH0 A0A0K8TS94 A0A1Y1KRT1 A0A2P8YFU0 A0A2M4AWS2 D6WX59 A0A067R0Z5 A0A2J7R3E6 A0A182RPE3 W5JWY4 A0A2M3ZDD6 A0A2M4AWU1 A0A182F719 A0A2M4C480 A0A182ULP0 A0A182JZ11 A0A182UGR0 A0A182XIS7 A0A182LBW9 Q7PR61 A0A182HTI9 T1DPZ6 Q9VEV5 A0A1Q3G4A9 A0A182VRU3 A0A3B0JQT7 Q295W3 B4GFL6 A0A084W348 A0A182QK22 A0A182LWT1 B4QX34 B4PQZ7 B4IBW0 B3P122 A0A182J0Z1 A0A1W4U7T6 B4KBZ6 A0A195C1U7 B0XG83 A0A023ED59 A0A182N7B7 B4LX41 A0A195AXS1 A0A182PE03 B3M2J7 A0A1I8NI56 Q0IFW2 B4NLA1 A0A195DB01 A0A1L8DMK6 A0A1I8PKG2 A0A0M4ERD4 K7IN17 J9JNG9 A0A158NLU9 A0A1L8EDI6 N6UGX2 A0A232F128 A0A1W4WRC6 A0A151X7Y8 A0A1B0CEE5 A0A195F4X4 B4JFL2 B5BV16 A0A034WWK1 A0A0C9QHW2 A0A0K8V4A6 A0A2H8TIU1 A0A0N0BJQ5 A0A0J7KYB7 E2AF96 A0A1A9WFN9 E9JBY1 A0A1B6DAT2 A0A0P6B4U6 A0A0P5Z7C1 A0A0A1X386 W8BZ50 A0A1B0G4T4 A0A1A9VD31 A0A0P5V7G9 A0A0L0BQI8 A0A1A9ZAD1 A0A0P5DZX8 A0A0P5T8U7 A0A1A9XBD3 A0A0P4WKQ5 A0A154PMU1
PDB
5AJ4
E-value=3.68317e-19,
Score=226
Ontologies
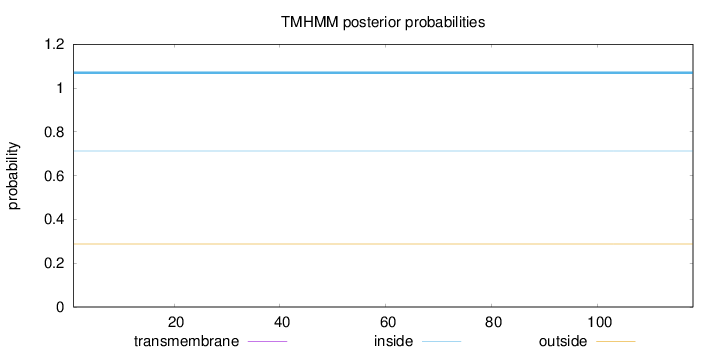
Topology
Length:
118
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00258
Exp number, first 60 AAs:
0.00253
Total prob of N-in:
0.71275
inside
1 - 118
Population Genetic Test Statistics
Pi
447.754846
Theta
213.700249
Tajima's D
3.417434
CLR
0.320667
CSRT
0.992150392480376
Interpretation
Uncertain
