Gene
KWMTBOMO10042
Pre Gene Modal
BGIBMGA005529
Annotation
PREDICTED:_N-sulphoglucosamine_sulphohydrolase_[Plutella_xylostella]
Full name
N-sulphoglucosamine sulphohydrolase
Alternative Name
Sulfoglucosamine sulfamidase
Sulphamidase
Sulphamidase
Location in the cell
Extracellular Reliability : 0.917
Sequence
CDS
ATGCGAGTTACCGGCATCAAACTTGCCCTTATCTTTTATTTTTTCATTACTGAAACTGTATTATCTGATAAGGTCCGCAATGTTCTTTTACTATTAGCGGACGATGGCGGTTTTGAGATAGGCGCGTATCGCAACAAAATATGTCAAACGCCGAATATAGATGAATTAGCGCGAAACGGCCTGTTGTTCAACAATGCATTCACATCAGTGAGCAGTTGCTCCCCAAGTCGGGCAGCACTCCTAACTGGGTCGCCGAGTCACCAGAATGGAATGTACGGGCTGCACCACGGCGTCCACCACTTCAACTCCTTCGACAACGTGACCAGTCTGCCGAACTTACTGCGCCAGAACGGAATTATGACGGGTATAATTGGTAAAAAGCACGTCGGGCCTTCGAGCGTGTATCAGTTTGATTATGAGCAGACGGAGGAGAACAACCACATTAATCAGGTCGGTCGCAACATCACCCACATGAAGCTGCTCGCCCGGGAGTTCATCGCGAGTGCAAACAAGGAAAACAAGCCCTTCTTCTTGTACGTGGCCTTCCATGACCCGCACCGCTGCGGACACAGCGACCCGCAGTACGGGCCCTTCTGCGAGAGGTTCGGCTCCGGCGAAGAAGGAATGGGCACCATTCCAGACTGGCAGCCGTGGTACTATCAGTGGGACGAGATTCAACTACCTTACTTCATACAGGACACTGAGGCGGCCAGGAGGGACATTGCCGCCCAGTACACAACGATGTCGCGACTCGACCAAGGGGTCGCCCTTATTCTGAAAGAACTGGAAAGCGCAGGGCACGCTGACGATACGTTAGTCATCTACACTTCAGACAACGGCATACCATTTCCATCGGGGAGAACTAATTTTTACGATCCGGGATTACGGGAGCCCCTCATCATTCGCTCACCTTCATCTTCAGCCAGAAAAAATGAGGCTTCAGGCGCCATGGTCAGCTTACTGGACATAATGCCAACCGTGCTAGACTGGTTTGGAATTGAGAAAGAAATGACAAATGACATTTGGGATGGAGATACACCAAAGAGTCTCTTACCTATACTGGAGAAAGAGCCGCCGCCCTCGGACTCGGACGCGGTGTTCGCCTCCCAGACGCACCACGAGATCACGATGTACTACCCGATGCGCGCCGTGCGCACGCGCCGCTACAAGCTGCTGCACAACCTCAACTACGGGATGCCGTTCCCGATCGACCAAGATCTATACGTCTCGCCCACATTTCAGGACCTATTAAACAGAACGCGCAGCAAGCAGCCTCTACCTTGGTACAAGACGCTGAAGCAATATTACTACCGGCCGCAGTGGGAGCTCTACGACCTTCGTAAGGATCCGGCAGAAACCAATAACTTACATGGTAAACCATCCCTGGAGGAGGTGGAGTCGAATCTCCGAGAACGATTGTCGCGATGGCAGCGCTCCACCCGCGACCCGTGGCTGTGTTCGCCCGACGCCGTCATGGAACGCGACTCTGACAACGCCGTGTGCCGCAGTCTAGACAACGGACTCCTAAACTATCATACACTATAG
Protein
MRVTGIKLALIFYFFITETVLSDKVRNVLLLLADDGGFEIGAYRNKICQTPNIDELARNGLLFNNAFTSVSSCSPSRAALLTGSPSHQNGMYGLHHGVHHFNSFDNVTSLPNLLRQNGIMTGIIGKKHVGPSSVYQFDYEQTEENNHINQVGRNITHMKLLAREFIASANKENKPFFLYVAFHDPHRCGHSDPQYGPFCERFGSGEEGMGTIPDWQPWYYQWDEIQLPYFIQDTEAARRDIAAQYTTMSRLDQGVALILKELESAGHADDTLVIYTSDNGIPFPSGRTNFYDPGLREPLIIRSPSSSARKNEASGAMVSLLDIMPTVLDWFGIEKEMTNDIWDGDTPKSLLPILEKEPPPSDSDAVFASQTHHEITMYYPMRAVRTRRYKLLHNLNYGMPFPIDQDLYVSPTFQDLLNRTRSKQPLPWYKTLKQYYYRPQWELYDLRKDPAETNNLHGKPSLEEVESNLRERLSRWQRSTRDPWLCSPDAVMERDSDNAVCRSLDNGLLNYHTL
Summary
Description
Catalyzes a step in lysosomal heparan sulfate degradation.
Catalytic Activity
H2O + N-sulfo-D-glucosamine = D-glucosamine + sulfate
Cofactor
Ca(2+)
Similarity
Belongs to the sulfatase family.
Keywords
3D-structure
Calcium
Complete proteome
Direct protein sequencing
Disease mutation
Disulfide bond
Glycoprotein
Hydrolase
Lysosome
Metal-binding
Mucopolysaccharidosis
Polymorphism
Reference proteome
Signal
Feature
chain N-sulphoglucosamine sulphohydrolase
sequence variant In MPS3A; dbSNP:rs139850991.
sequence variant In MPS3A; dbSNP:rs139850991.
Uniprot
A0A3S2NJ57
H9J7N7
A0A0N1PJL8
A0A2A4JJH2
A0A212FF84
A0A2W1BLH5
+ More
A0A232FA88 K7J4V3 A0A336LBG8 A0A026WG24 A0A3L8DFF8 D6WXJ3 A0A0L7R4P9 A0A336KLZ7 A0A154P5W2 E2A6X9 A0A151X7X6 A0A088A242 A0A0M8ZZG7 A0A1B0D306 A0A1Y1LQD7 A0A067R9H1 A0A2J7RAV0 E2BZ75 F4W713 A0A151K0T4 A0A195C3R2 A0A182NSP7 A0A158NMB5 N6U038 A0A182XG83 A0A182W1E0 W5JQ78 A0A182PLV8 A0A1J1J0M6 A0A1S4FQA7 A0A182L776 A0A182IKU3 Q7PH12 A0A182GEZ8 A0A182IDC8 A0A2A3E146 A0A182KHP6 A0A182Q2U5 A0A084VNQ6 A0A0N8ES60 A0A1Q3FX62 A0A182U4Y7 A0A195DB10 A0A0J7K6A0 B4JGE7 A0A182RZJ1 A0A0K8W479 A0A034WT68 W8C7R9 Q16T02 A0A182G033 T1PDX4 A0A182Y5G7 A0A0A1WE65 A0A0L0CJH2 B3MSR6 A0A182SY23 A0A3B0KG69 Q298E8 A0A1B6EZN8 A0A182VGY4 B4G3P1 A0A182MJV8 F1NGI6 A0A293MCB0 B4K587 A0A226N1E7 A0A1S3HRU1 A0A1S3ILX1 A0A2K6FXZ7 B4PM30 A0A340XTI2 A0A3L8S0V7 A0A2K5HNE5 F6RC31 A0A1I8P9M3 A0A2R8ZKN1 A0A096NTB4 A0A218UL41 G3RBT8 A0A2Y9FM96 A0A2B4SGR8 Q8I1A5 P51688 A0A093GEM9 A0A2K5S515 A0A2K6SR45 A0A2K5C3H8 A0A1U7SZX2 H0V4W6 A0A2K6B4C9 F7DFZ7 A0A341CR07 U6CZ92
A0A232FA88 K7J4V3 A0A336LBG8 A0A026WG24 A0A3L8DFF8 D6WXJ3 A0A0L7R4P9 A0A336KLZ7 A0A154P5W2 E2A6X9 A0A151X7X6 A0A088A242 A0A0M8ZZG7 A0A1B0D306 A0A1Y1LQD7 A0A067R9H1 A0A2J7RAV0 E2BZ75 F4W713 A0A151K0T4 A0A195C3R2 A0A182NSP7 A0A158NMB5 N6U038 A0A182XG83 A0A182W1E0 W5JQ78 A0A182PLV8 A0A1J1J0M6 A0A1S4FQA7 A0A182L776 A0A182IKU3 Q7PH12 A0A182GEZ8 A0A182IDC8 A0A2A3E146 A0A182KHP6 A0A182Q2U5 A0A084VNQ6 A0A0N8ES60 A0A1Q3FX62 A0A182U4Y7 A0A195DB10 A0A0J7K6A0 B4JGE7 A0A182RZJ1 A0A0K8W479 A0A034WT68 W8C7R9 Q16T02 A0A182G033 T1PDX4 A0A182Y5G7 A0A0A1WE65 A0A0L0CJH2 B3MSR6 A0A182SY23 A0A3B0KG69 Q298E8 A0A1B6EZN8 A0A182VGY4 B4G3P1 A0A182MJV8 F1NGI6 A0A293MCB0 B4K587 A0A226N1E7 A0A1S3HRU1 A0A1S3ILX1 A0A2K6FXZ7 B4PM30 A0A340XTI2 A0A3L8S0V7 A0A2K5HNE5 F6RC31 A0A1I8P9M3 A0A2R8ZKN1 A0A096NTB4 A0A218UL41 G3RBT8 A0A2Y9FM96 A0A2B4SGR8 Q8I1A5 P51688 A0A093GEM9 A0A2K5S515 A0A2K6SR45 A0A2K5C3H8 A0A1U7SZX2 H0V4W6 A0A2K6B4C9 F7DFZ7 A0A341CR07 U6CZ92
EC Number
3.10.1.1
Pubmed
19121390
26354079
22118469
28756777
28648823
20075255
+ More
24508170 30249741 18362917 19820115 20798317 28004739 24845553 21719571 21347285 23537049 20920257 23761445 20966253 12364791 14747013 17210077 26483478 24438588 26999592 17994087 25348373 24495485 17510324 25315136 25244985 25830018 26108605 15632085 15592404 17550304 30282656 19892987 22722832 22398555 18057021 7493035 14702039 15489334 12754519 19159218 24275569 24816101 9158154 9285796 9401012 9554748 9744479 11182930 12000360 11793481 12702166 15146460 15637719 15902564 17128482 16311287 18407553 21671382 28101780 21993624 17495919
24508170 30249741 18362917 19820115 20798317 28004739 24845553 21719571 21347285 23537049 20920257 23761445 20966253 12364791 14747013 17210077 26483478 24438588 26999592 17994087 25348373 24495485 17510324 25315136 25244985 25830018 26108605 15632085 15592404 17550304 30282656 19892987 22722832 22398555 18057021 7493035 14702039 15489334 12754519 19159218 24275569 24816101 9158154 9285796 9401012 9554748 9744479 11182930 12000360 11793481 12702166 15146460 15637719 15902564 17128482 16311287 18407553 21671382 28101780 21993624 17495919
EMBL
RSAL01000075
RVE48897.1
BABH01020426
KQ460044
KPJ18297.1
NWSH01001312
+ More
PCG71744.1 AGBW02008849 OWR52392.1 KZ150179 PZC72533.1 NNAY01000614 OXU27390.1 AAZX01007073 UFQS01001502 UFQT01001502 SSX11284.1 SSX30852.1 KK107231 EZA55070.1 QOIP01000009 RLU18932.1 KQ971361 EFA08854.1 KQ414657 KOC65833.1 UFQS01000253 UFQT01000253 SSX02060.1 SSX22437.1 KQ434823 KZC07309.1 GL437203 EFN70845.1 KQ982431 KYQ56472.1 KQ435812 KOX72773.1 AJVK01010815 GEZM01051103 JAV75171.1 KK852804 KDR16259.1 NEVH01006564 PNF37952.1 GL451577 EFN78981.1 GL887802 EGI70003.1 KQ981253 KYN44176.1 KQ978344 KYM94821.1 ADTU01002232 ADTU01002233 APGK01047787 KB741089 ENN73961.1 ADMH02000787 ETN65055.1 CVRI01000064 CRL04974.1 AAAB01008880 EAA44728.2 JXUM01058904 KQ562024 KXJ76879.1 APCN01005106 KZ288467 PBC25417.1 AXCN02000231 ATLV01014835 KE524991 KFB39600.1 GDUN01000370 JAN95549.1 GFDL01002878 JAV32167.1 KQ981042 KYN10073.1 LBMM01012961 KMQ85897.1 CH916369 EDV92616.1 GDHF01006634 JAI45680.1 GAKP01001567 JAC57385.1 GAMC01000367 JAC06189.1 CH477662 EAT37588.1 KA646138 AFP60767.1 GBXI01017311 JAC96980.1 JRES01000310 KNC32385.1 CH902623 EDV30306.1 OUUW01000007 SPP82638.1 CM000070 EAL28007.2 GECZ01026391 JAS43378.1 CH479179 EDW24362.1 AXCM01006741 AADN05000628 GFWV01013723 MAA38452.1 CH933806 EDW15087.1 MCFN01000295 OXB61069.1 CM000160 EDW95962.1 QUSF01000104 RLV92050.1 AJFE02110123 AJFE02110124 AHZZ02010799 MUZQ01000239 OWK54489.1 CABD030035768 CABD030035769 LSMT01000096 PFX27757.1 AY190951 CH964251 AAO01067.1 EDW83222.2 U30894 U60111 U60107 U60108 U60109 U60110 AK291257 BC047318 KL215558 KFV65337.1 AAKN02047339 HAAF01004165 CCP75990.1
PCG71744.1 AGBW02008849 OWR52392.1 KZ150179 PZC72533.1 NNAY01000614 OXU27390.1 AAZX01007073 UFQS01001502 UFQT01001502 SSX11284.1 SSX30852.1 KK107231 EZA55070.1 QOIP01000009 RLU18932.1 KQ971361 EFA08854.1 KQ414657 KOC65833.1 UFQS01000253 UFQT01000253 SSX02060.1 SSX22437.1 KQ434823 KZC07309.1 GL437203 EFN70845.1 KQ982431 KYQ56472.1 KQ435812 KOX72773.1 AJVK01010815 GEZM01051103 JAV75171.1 KK852804 KDR16259.1 NEVH01006564 PNF37952.1 GL451577 EFN78981.1 GL887802 EGI70003.1 KQ981253 KYN44176.1 KQ978344 KYM94821.1 ADTU01002232 ADTU01002233 APGK01047787 KB741089 ENN73961.1 ADMH02000787 ETN65055.1 CVRI01000064 CRL04974.1 AAAB01008880 EAA44728.2 JXUM01058904 KQ562024 KXJ76879.1 APCN01005106 KZ288467 PBC25417.1 AXCN02000231 ATLV01014835 KE524991 KFB39600.1 GDUN01000370 JAN95549.1 GFDL01002878 JAV32167.1 KQ981042 KYN10073.1 LBMM01012961 KMQ85897.1 CH916369 EDV92616.1 GDHF01006634 JAI45680.1 GAKP01001567 JAC57385.1 GAMC01000367 JAC06189.1 CH477662 EAT37588.1 KA646138 AFP60767.1 GBXI01017311 JAC96980.1 JRES01000310 KNC32385.1 CH902623 EDV30306.1 OUUW01000007 SPP82638.1 CM000070 EAL28007.2 GECZ01026391 JAS43378.1 CH479179 EDW24362.1 AXCM01006741 AADN05000628 GFWV01013723 MAA38452.1 CH933806 EDW15087.1 MCFN01000295 OXB61069.1 CM000160 EDW95962.1 QUSF01000104 RLV92050.1 AJFE02110123 AJFE02110124 AHZZ02010799 MUZQ01000239 OWK54489.1 CABD030035768 CABD030035769 LSMT01000096 PFX27757.1 AY190951 CH964251 AAO01067.1 EDW83222.2 U30894 U60111 U60107 U60108 U60109 U60110 AK291257 BC047318 KL215558 KFV65337.1 AAKN02047339 HAAF01004165 CCP75990.1
Proteomes
UP000283053
UP000005204
UP000053240
UP000218220
UP000007151
UP000215335
+ More
UP000002358 UP000053097 UP000279307 UP000007266 UP000053825 UP000076502 UP000000311 UP000075809 UP000005203 UP000053105 UP000092462 UP000027135 UP000235965 UP000008237 UP000007755 UP000078541 UP000078542 UP000075884 UP000005205 UP000019118 UP000076407 UP000075920 UP000000673 UP000075885 UP000183832 UP000075882 UP000075880 UP000007062 UP000069940 UP000249989 UP000075840 UP000242457 UP000075881 UP000075886 UP000030765 UP000075902 UP000078492 UP000036403 UP000001070 UP000075900 UP000008820 UP000069272 UP000095301 UP000076408 UP000037069 UP000007801 UP000075901 UP000268350 UP000001819 UP000075903 UP000008744 UP000075883 UP000000539 UP000009192 UP000198323 UP000085678 UP000233160 UP000002282 UP000265300 UP000276834 UP000233080 UP000002281 UP000095300 UP000240080 UP000028761 UP000197619 UP000001519 UP000248484 UP000225706 UP000007798 UP000005640 UP000053875 UP000233040 UP000233220 UP000233020 UP000189704 UP000005447 UP000233120 UP000002280 UP000252040
UP000002358 UP000053097 UP000279307 UP000007266 UP000053825 UP000076502 UP000000311 UP000075809 UP000005203 UP000053105 UP000092462 UP000027135 UP000235965 UP000008237 UP000007755 UP000078541 UP000078542 UP000075884 UP000005205 UP000019118 UP000076407 UP000075920 UP000000673 UP000075885 UP000183832 UP000075882 UP000075880 UP000007062 UP000069940 UP000249989 UP000075840 UP000242457 UP000075881 UP000075886 UP000030765 UP000075902 UP000078492 UP000036403 UP000001070 UP000075900 UP000008820 UP000069272 UP000095301 UP000076408 UP000037069 UP000007801 UP000075901 UP000268350 UP000001819 UP000075903 UP000008744 UP000075883 UP000000539 UP000009192 UP000198323 UP000085678 UP000233160 UP000002282 UP000265300 UP000276834 UP000233080 UP000002281 UP000095300 UP000240080 UP000028761 UP000197619 UP000001519 UP000248484 UP000225706 UP000007798 UP000005640 UP000053875 UP000233040 UP000233220 UP000233020 UP000189704 UP000005447 UP000233120 UP000002280 UP000252040
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A3S2NJ57
H9J7N7
A0A0N1PJL8
A0A2A4JJH2
A0A212FF84
A0A2W1BLH5
+ More
A0A232FA88 K7J4V3 A0A336LBG8 A0A026WG24 A0A3L8DFF8 D6WXJ3 A0A0L7R4P9 A0A336KLZ7 A0A154P5W2 E2A6X9 A0A151X7X6 A0A088A242 A0A0M8ZZG7 A0A1B0D306 A0A1Y1LQD7 A0A067R9H1 A0A2J7RAV0 E2BZ75 F4W713 A0A151K0T4 A0A195C3R2 A0A182NSP7 A0A158NMB5 N6U038 A0A182XG83 A0A182W1E0 W5JQ78 A0A182PLV8 A0A1J1J0M6 A0A1S4FQA7 A0A182L776 A0A182IKU3 Q7PH12 A0A182GEZ8 A0A182IDC8 A0A2A3E146 A0A182KHP6 A0A182Q2U5 A0A084VNQ6 A0A0N8ES60 A0A1Q3FX62 A0A182U4Y7 A0A195DB10 A0A0J7K6A0 B4JGE7 A0A182RZJ1 A0A0K8W479 A0A034WT68 W8C7R9 Q16T02 A0A182G033 T1PDX4 A0A182Y5G7 A0A0A1WE65 A0A0L0CJH2 B3MSR6 A0A182SY23 A0A3B0KG69 Q298E8 A0A1B6EZN8 A0A182VGY4 B4G3P1 A0A182MJV8 F1NGI6 A0A293MCB0 B4K587 A0A226N1E7 A0A1S3HRU1 A0A1S3ILX1 A0A2K6FXZ7 B4PM30 A0A340XTI2 A0A3L8S0V7 A0A2K5HNE5 F6RC31 A0A1I8P9M3 A0A2R8ZKN1 A0A096NTB4 A0A218UL41 G3RBT8 A0A2Y9FM96 A0A2B4SGR8 Q8I1A5 P51688 A0A093GEM9 A0A2K5S515 A0A2K6SR45 A0A2K5C3H8 A0A1U7SZX2 H0V4W6 A0A2K6B4C9 F7DFZ7 A0A341CR07 U6CZ92
A0A232FA88 K7J4V3 A0A336LBG8 A0A026WG24 A0A3L8DFF8 D6WXJ3 A0A0L7R4P9 A0A336KLZ7 A0A154P5W2 E2A6X9 A0A151X7X6 A0A088A242 A0A0M8ZZG7 A0A1B0D306 A0A1Y1LQD7 A0A067R9H1 A0A2J7RAV0 E2BZ75 F4W713 A0A151K0T4 A0A195C3R2 A0A182NSP7 A0A158NMB5 N6U038 A0A182XG83 A0A182W1E0 W5JQ78 A0A182PLV8 A0A1J1J0M6 A0A1S4FQA7 A0A182L776 A0A182IKU3 Q7PH12 A0A182GEZ8 A0A182IDC8 A0A2A3E146 A0A182KHP6 A0A182Q2U5 A0A084VNQ6 A0A0N8ES60 A0A1Q3FX62 A0A182U4Y7 A0A195DB10 A0A0J7K6A0 B4JGE7 A0A182RZJ1 A0A0K8W479 A0A034WT68 W8C7R9 Q16T02 A0A182G033 T1PDX4 A0A182Y5G7 A0A0A1WE65 A0A0L0CJH2 B3MSR6 A0A182SY23 A0A3B0KG69 Q298E8 A0A1B6EZN8 A0A182VGY4 B4G3P1 A0A182MJV8 F1NGI6 A0A293MCB0 B4K587 A0A226N1E7 A0A1S3HRU1 A0A1S3ILX1 A0A2K6FXZ7 B4PM30 A0A340XTI2 A0A3L8S0V7 A0A2K5HNE5 F6RC31 A0A1I8P9M3 A0A2R8ZKN1 A0A096NTB4 A0A218UL41 G3RBT8 A0A2Y9FM96 A0A2B4SGR8 Q8I1A5 P51688 A0A093GEM9 A0A2K5S515 A0A2K6SR45 A0A2K5C3H8 A0A1U7SZX2 H0V4W6 A0A2K6B4C9 F7DFZ7 A0A341CR07 U6CZ92
PDB
4MIV
E-value=2.20617e-165,
Score=1495
Ontologies
PATHWAY
GO
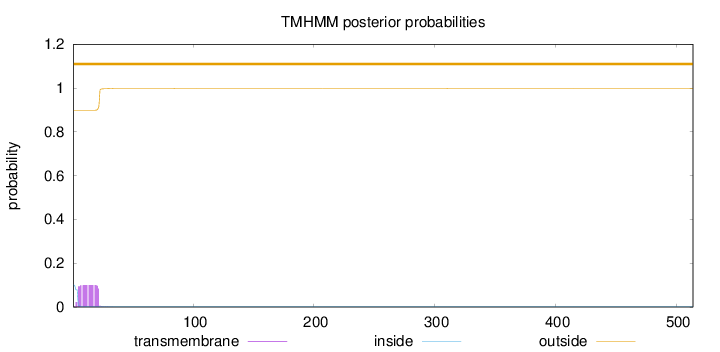
Topology
Subcellular location
Lysosome
SignalP
Position: 1 - 22,
Likelihood: 0.930554
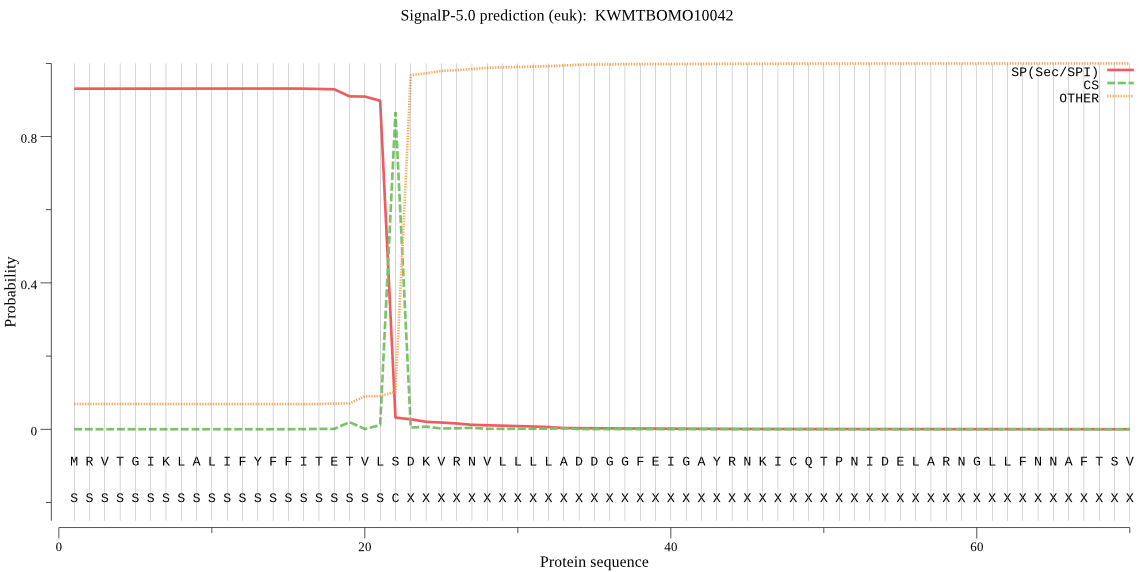
Length:
514
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.83414
Exp number, first 60 AAs:
1.83087
Total prob of N-in:
0.10134
outside
1 - 514
Population Genetic Test Statistics
Pi
199.054881
Theta
147.20731
Tajima's D
0.137092
CLR
0.289695
CSRT
0.410029498525074
Interpretation
Uncertain
