Gene
KWMTBOMO10041 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005687
Annotation
Phosphomannomutase_[Operophtera_brumata]
Full name
Phosphomannomutase
Location in the cell
Cytoplasmic Reliability : 2.123 Nuclear Reliability : 2.215
Sequence
CDS
ATGACTAGTCAAAAGAAAGTTTTGTATTTGTTCGATGTTGACGGAACGTTGACAAAACCGCGCCAAAAAATTACAGAAGAATTCAGGAGGTTCATTCTTGACGAGGTAAAATCAAAAGTGGATGTAGGATTGGTCAGTGGTTCAGATTATATGAAGATTTCCGAACAAATGGGCGGAGAGGACGTCGTGTCCAATTTCAACTACGTATTCAGTGAAAATGGACTGGTGCATCATAAGAACGGAAAAAAATTAAGTTCAGAGAGCATAGTTAACCATTTGGGTGAACAAAAACTGCAAGAGGTAATAAACTTCGCCATGGGTTACATGTCCAACATCAAATTACCAGTCAAGAGAGGTAATTTCATTGAATTCCGATCAAGTATGCTTAATATTTGCCCTGTTGGAAGATCATGTAATCAAATAGAGCGAGATCAGTTTTCTGAGTATGATTCAAAACACAAAGTTAGGCACCAATTTGTTGAAGCACTTCAAAGCAAGTTCAAAGACTCGGGTTTAAAGTTTGCTCTAGGAGGTCAAATCAGTATTGATGTGTTCCCAATTGGATGGGACAAGACATACTGCTTAAATCACATAGCTAGTGAGAATTTTGCTGAAATTCACTTCTTTGGAGACAAAACAGACCCTGGTGGGAATGATCATGAAATTTACAATGACTCTAGGACTGTGGGACACAGAGTCTCATCTCCAAGTGACACTGAAATGCAAGTAAAAAAATGTCTTAATGTGGAATAG
Protein
MTSQKKVLYLFDVDGTLTKPRQKITEEFRRFILDEVKSKVDVGLVSGSDYMKISEQMGGEDVVSNFNYVFSENGLVHHKNGKKLSSESIVNHLGEQKLQEVINFAMGYMSNIKLPVKRGNFIEFRSSMLNICPVGRSCNQIERDQFSEYDSKHKVRHQFVEALQSKFKDSGLKFALGGQISIDVFPIGWDKTYCLNHIASENFAEIHFFGDKTDPGGNDHEIYNDSRTVGHRVSSPSDTEMQVKKCLNVE
Summary
Description
Involved in the synthesis of the GDP-mannose and dolichol-phosphate-mannose required for a number of critical mannosyl transfer reactions.
Catalytic Activity
alpha-D-mannose 1-phosphate = D-mannose 6-phosphate
Subunit
Homodimer.
Similarity
Belongs to the eukaryotic PMM family.
Uniprot
H9J845
A0A2W1BB19
A0A2A4JJZ2
A0A0L7LSL4
S4PMU4
I4DK74
+ More
A0A2H1WVB5 A0A2I9LPG3 A0A1S3HCV3 A0A1S3KEL4 E9HCF1 A0A1B6DIM5 A0A0P4WB28 A0A2P8YKY2 V4BAW4 C3YWF0 D6WXJ2 A0A0P6BS41 A0A0P5R453 A0A2J7RAT9 A0A2J7RAU5 A0A0T6AZ43 A0A1B6G4M1 A0A1B6M126 A0A210QWA3 H2YEQ1 A0A1W4WZA5 A0A1V9XHX8 A0A2T7PYN3 A0A1B6IJL6 A0A1Y1LQR5 J9JYE1 A0A2P2I7N5 A0A0P6HUS4 A0A369RZY6 B3RMY2 C4WU27 A0A087UM49 A0A1Q3F690 F6SS61 A0A1W2WPB5 B0WHH5 A0A0K8R750 V5GG89 J3JVF0 G1KD11 A0A131XZ82 A0A023FY01 A0A0C9S225 A0A0P4VUS9 R4FJZ3 A0A1E1X9L8 T1KR88 B7QIG2 H2THT1 U5EKG9 A0A131XDM1 V8P3L0 A0A023GGU3 H9H6Z7 G3MPQ8 A0A131YIZ8 A0A182GCZ4 A0A1W4UVF1 A0A212FF79 T1D9U4 A0A224Z5T5 A0A023FHX0 A0A1D5PQW2 E2A6X8 H2RJ98 A0A023ELB7 A0A195C1U8 A0A151NHF5 A0A1J1IHA7 A0A182GNB4 A0A1V4JB57 A0A0N0PDS2 A0A1W7REV7 A0A0F7Z640 Q0IEM8 A0A183QQP1 A0A218UZV2 A0A293LZ40 Q6GNE0 J3S998 U3FWE0 A0A2P4W7H3 A0A194QAS7 A0A2P6K547 A0A182LXJ3 G1NY02 A0A1B0CL90 A0A3Q7RBJ5 A0A2Y9KEQ3 H0ZHY9 A0A1L8GGM9 A0A2D4M6K7 A0A287D5V8 A0A1S4FMN1
A0A2H1WVB5 A0A2I9LPG3 A0A1S3HCV3 A0A1S3KEL4 E9HCF1 A0A1B6DIM5 A0A0P4WB28 A0A2P8YKY2 V4BAW4 C3YWF0 D6WXJ2 A0A0P6BS41 A0A0P5R453 A0A2J7RAT9 A0A2J7RAU5 A0A0T6AZ43 A0A1B6G4M1 A0A1B6M126 A0A210QWA3 H2YEQ1 A0A1W4WZA5 A0A1V9XHX8 A0A2T7PYN3 A0A1B6IJL6 A0A1Y1LQR5 J9JYE1 A0A2P2I7N5 A0A0P6HUS4 A0A369RZY6 B3RMY2 C4WU27 A0A087UM49 A0A1Q3F690 F6SS61 A0A1W2WPB5 B0WHH5 A0A0K8R750 V5GG89 J3JVF0 G1KD11 A0A131XZ82 A0A023FY01 A0A0C9S225 A0A0P4VUS9 R4FJZ3 A0A1E1X9L8 T1KR88 B7QIG2 H2THT1 U5EKG9 A0A131XDM1 V8P3L0 A0A023GGU3 H9H6Z7 G3MPQ8 A0A131YIZ8 A0A182GCZ4 A0A1W4UVF1 A0A212FF79 T1D9U4 A0A224Z5T5 A0A023FHX0 A0A1D5PQW2 E2A6X8 H2RJ98 A0A023ELB7 A0A195C1U8 A0A151NHF5 A0A1J1IHA7 A0A182GNB4 A0A1V4JB57 A0A0N0PDS2 A0A1W7REV7 A0A0F7Z640 Q0IEM8 A0A183QQP1 A0A218UZV2 A0A293LZ40 Q6GNE0 J3S998 U3FWE0 A0A2P4W7H3 A0A194QAS7 A0A2P6K547 A0A182LXJ3 G1NY02 A0A1B0CL90 A0A3Q7RBJ5 A0A2Y9KEQ3 H0ZHY9 A0A1L8GGM9 A0A2D4M6K7 A0A287D5V8 A0A1S4FMN1
EC Number
5.4.2.8
Pubmed
19121390
28756777
26227816
23622113
22651552
29248469
+ More
21292972 29403074 23254933 18563158 18362917 19820115 28812685 28327890 28004739 30042472 18719581 12481130 15114417 22516182 23537049 26131772 27129103 28503490 21551351 28049606 24297900 17495919 22216098 26830274 26483478 22118469 23758969 28797301 15592404 20798317 24945155 22293439 26354079 26358130 17510324 23025625 23915248 26114425 26394399 21993624 20360741 27762356
21292972 29403074 23254933 18563158 18362917 19820115 28812685 28327890 28004739 30042472 18719581 12481130 15114417 22516182 23537049 26131772 27129103 28503490 21551351 28049606 24297900 17495919 22216098 26830274 26483478 22118469 23758969 28797301 15592404 20798317 24945155 22293439 26354079 26358130 17510324 23025625 23915248 26114425 26394399 21993624 20360741 27762356
EMBL
BABH01020426
KZ150179
PZC72532.1
NWSH01001312
PCG71743.1
JTDY01000229
+ More
KOB78176.1 GAIX01003375 JAA89185.1 AK401692 BAM18314.1 ODYU01011325 SOQ57001.1 GFWZ01000283 MBW20273.1 GL732619 EFX70619.1 GEDC01011747 JAS25551.1 GDRN01067174 GDRN01067172 GDRN01067171 GDRN01067169 JAI64412.1 PYGN01000522 PSN44908.1 KB200109 ESP03112.1 GG666561 EEN55194.1 KQ971361 EFA07975.2 GDIP01010451 JAM93264.1 GDIQ01203822 GDIQ01105748 JAL45978.1 NEVH01006564 PNF37953.1 PNF37954.1 LJIG01022525 KRT80117.1 GECZ01012393 JAS57376.1 GEBQ01010341 JAT29636.1 NEDP02001549 OWF52993.1 MNPL01010405 OQR73140.1 PZQS01000001 PVD38507.1 GECU01020595 JAS87111.1 GEZM01051111 GEZM01051110 GEZM01051109 GEZM01051107 GEZM01051105 GEZM01051104 JAV75168.1 ABLF02026171 IACF01004451 LAB70044.1 GDIQ01014638 JAN80099.1 NOWV01000119 RDD39851.1 DS985242 EDV27355.1 AK340880 BAH71397.1 KK120523 KFM78438.1 GFDL01011960 JAV23085.1 EAAA01001620 DS231936 EDS27753.1 GADI01006823 JAA66985.1 GALX01005492 JAB62974.1 APGK01047787 BT127218 KB741089 KB632382 AEE62180.1 ENN73958.1 ERL94187.1 GEFM01003477 JAP72319.1 GBBL01001717 JAC25603.1 GBZX01001766 JAG90974.1 GDKW01002025 JAI54570.1 GAHY01002083 JAA75427.1 GFAC01003482 JAT95706.1 CAEY01000384 ABJB010713827 DS945857 EEC18634.1 GANO01001817 JAB58054.1 GEFH01004373 JAP64208.1 AZIM01000983 ETE68593.1 GBBM01002207 JAC33211.1 JO843859 AEO35476.1 GEDV01010656 JAP77901.1 JXUM01054927 KQ561844 KXJ77367.1 AGBW02008849 OWR52391.1 GAAZ01001789 JAA96154.1 GFPF01010608 MAA21754.1 GBBK01003430 JAC21052.1 AADN05000523 GL437203 EFN70844.1 GAPW01003592 JAC10006.1 KQ978344 KYM94822.1 AKHW03002956 KYO36242.1 CVRI01000047 CRK98422.1 JXUM01076094 KQ562924 KXJ74824.1 LSYS01008398 OPJ68987.1 KQ460044 KPJ18298.1 GDAY02001766 JAV49662.1 GBEX01002489 JAI12071.1 CH477586 EAT38617.1 MUZQ01000080 OWK59283.1 GFWV01009244 MAA33973.1 BC073573 AAH73573.1 JU175414 AFJ50938.1 GAEP01001135 JAB53686.1 LFJK02000008 POM45458.1 KQ459232 KPJ02534.1 MWRG01026311 PRD21448.1 AXCM01001158 AAPE02036788 AJWK01017046 ABQF01015865 CM004473 OCT82987.1 IACM01081533 LAB29017.1 AGTP01024032
KOB78176.1 GAIX01003375 JAA89185.1 AK401692 BAM18314.1 ODYU01011325 SOQ57001.1 GFWZ01000283 MBW20273.1 GL732619 EFX70619.1 GEDC01011747 JAS25551.1 GDRN01067174 GDRN01067172 GDRN01067171 GDRN01067169 JAI64412.1 PYGN01000522 PSN44908.1 KB200109 ESP03112.1 GG666561 EEN55194.1 KQ971361 EFA07975.2 GDIP01010451 JAM93264.1 GDIQ01203822 GDIQ01105748 JAL45978.1 NEVH01006564 PNF37953.1 PNF37954.1 LJIG01022525 KRT80117.1 GECZ01012393 JAS57376.1 GEBQ01010341 JAT29636.1 NEDP02001549 OWF52993.1 MNPL01010405 OQR73140.1 PZQS01000001 PVD38507.1 GECU01020595 JAS87111.1 GEZM01051111 GEZM01051110 GEZM01051109 GEZM01051107 GEZM01051105 GEZM01051104 JAV75168.1 ABLF02026171 IACF01004451 LAB70044.1 GDIQ01014638 JAN80099.1 NOWV01000119 RDD39851.1 DS985242 EDV27355.1 AK340880 BAH71397.1 KK120523 KFM78438.1 GFDL01011960 JAV23085.1 EAAA01001620 DS231936 EDS27753.1 GADI01006823 JAA66985.1 GALX01005492 JAB62974.1 APGK01047787 BT127218 KB741089 KB632382 AEE62180.1 ENN73958.1 ERL94187.1 GEFM01003477 JAP72319.1 GBBL01001717 JAC25603.1 GBZX01001766 JAG90974.1 GDKW01002025 JAI54570.1 GAHY01002083 JAA75427.1 GFAC01003482 JAT95706.1 CAEY01000384 ABJB010713827 DS945857 EEC18634.1 GANO01001817 JAB58054.1 GEFH01004373 JAP64208.1 AZIM01000983 ETE68593.1 GBBM01002207 JAC33211.1 JO843859 AEO35476.1 GEDV01010656 JAP77901.1 JXUM01054927 KQ561844 KXJ77367.1 AGBW02008849 OWR52391.1 GAAZ01001789 JAA96154.1 GFPF01010608 MAA21754.1 GBBK01003430 JAC21052.1 AADN05000523 GL437203 EFN70844.1 GAPW01003592 JAC10006.1 KQ978344 KYM94822.1 AKHW03002956 KYO36242.1 CVRI01000047 CRK98422.1 JXUM01076094 KQ562924 KXJ74824.1 LSYS01008398 OPJ68987.1 KQ460044 KPJ18298.1 GDAY02001766 JAV49662.1 GBEX01002489 JAI12071.1 CH477586 EAT38617.1 MUZQ01000080 OWK59283.1 GFWV01009244 MAA33973.1 BC073573 AAH73573.1 JU175414 AFJ50938.1 GAEP01001135 JAB53686.1 LFJK02000008 POM45458.1 KQ459232 KPJ02534.1 MWRG01026311 PRD21448.1 AXCM01001158 AAPE02036788 AJWK01017046 ABQF01015865 CM004473 OCT82987.1 IACM01081533 LAB29017.1 AGTP01024032
Proteomes
UP000005204
UP000218220
UP000037510
UP000085678
UP000000305
UP000245037
+ More
UP000030746 UP000001554 UP000007266 UP000235965 UP000242188 UP000007875 UP000192223 UP000192247 UP000245119 UP000007819 UP000253843 UP000009022 UP000054359 UP000008144 UP000002320 UP000019118 UP000030742 UP000001646 UP000015104 UP000001555 UP000005226 UP000002280 UP000069940 UP000249989 UP000192221 UP000007151 UP000000539 UP000000311 UP000078542 UP000050525 UP000183832 UP000190648 UP000053240 UP000008820 UP000050792 UP000197619 UP000237256 UP000053268 UP000075883 UP000001074 UP000092461 UP000286641 UP000248482 UP000007754 UP000186698 UP000005215
UP000030746 UP000001554 UP000007266 UP000235965 UP000242188 UP000007875 UP000192223 UP000192247 UP000245119 UP000007819 UP000253843 UP000009022 UP000054359 UP000008144 UP000002320 UP000019118 UP000030742 UP000001646 UP000015104 UP000001555 UP000005226 UP000002280 UP000069940 UP000249989 UP000192221 UP000007151 UP000000539 UP000000311 UP000078542 UP000050525 UP000183832 UP000190648 UP000053240 UP000008820 UP000050792 UP000197619 UP000237256 UP000053268 UP000075883 UP000001074 UP000092461 UP000286641 UP000248482 UP000007754 UP000186698 UP000005215
PRIDE
Pfam
PF03332 PMM
SUPFAM
SSF56784
SSF56784
Gene 3D
CDD
ProteinModelPortal
H9J845
A0A2W1BB19
A0A2A4JJZ2
A0A0L7LSL4
S4PMU4
I4DK74
+ More
A0A2H1WVB5 A0A2I9LPG3 A0A1S3HCV3 A0A1S3KEL4 E9HCF1 A0A1B6DIM5 A0A0P4WB28 A0A2P8YKY2 V4BAW4 C3YWF0 D6WXJ2 A0A0P6BS41 A0A0P5R453 A0A2J7RAT9 A0A2J7RAU5 A0A0T6AZ43 A0A1B6G4M1 A0A1B6M126 A0A210QWA3 H2YEQ1 A0A1W4WZA5 A0A1V9XHX8 A0A2T7PYN3 A0A1B6IJL6 A0A1Y1LQR5 J9JYE1 A0A2P2I7N5 A0A0P6HUS4 A0A369RZY6 B3RMY2 C4WU27 A0A087UM49 A0A1Q3F690 F6SS61 A0A1W2WPB5 B0WHH5 A0A0K8R750 V5GG89 J3JVF0 G1KD11 A0A131XZ82 A0A023FY01 A0A0C9S225 A0A0P4VUS9 R4FJZ3 A0A1E1X9L8 T1KR88 B7QIG2 H2THT1 U5EKG9 A0A131XDM1 V8P3L0 A0A023GGU3 H9H6Z7 G3MPQ8 A0A131YIZ8 A0A182GCZ4 A0A1W4UVF1 A0A212FF79 T1D9U4 A0A224Z5T5 A0A023FHX0 A0A1D5PQW2 E2A6X8 H2RJ98 A0A023ELB7 A0A195C1U8 A0A151NHF5 A0A1J1IHA7 A0A182GNB4 A0A1V4JB57 A0A0N0PDS2 A0A1W7REV7 A0A0F7Z640 Q0IEM8 A0A183QQP1 A0A218UZV2 A0A293LZ40 Q6GNE0 J3S998 U3FWE0 A0A2P4W7H3 A0A194QAS7 A0A2P6K547 A0A182LXJ3 G1NY02 A0A1B0CL90 A0A3Q7RBJ5 A0A2Y9KEQ3 H0ZHY9 A0A1L8GGM9 A0A2D4M6K7 A0A287D5V8 A0A1S4FMN1
A0A2H1WVB5 A0A2I9LPG3 A0A1S3HCV3 A0A1S3KEL4 E9HCF1 A0A1B6DIM5 A0A0P4WB28 A0A2P8YKY2 V4BAW4 C3YWF0 D6WXJ2 A0A0P6BS41 A0A0P5R453 A0A2J7RAT9 A0A2J7RAU5 A0A0T6AZ43 A0A1B6G4M1 A0A1B6M126 A0A210QWA3 H2YEQ1 A0A1W4WZA5 A0A1V9XHX8 A0A2T7PYN3 A0A1B6IJL6 A0A1Y1LQR5 J9JYE1 A0A2P2I7N5 A0A0P6HUS4 A0A369RZY6 B3RMY2 C4WU27 A0A087UM49 A0A1Q3F690 F6SS61 A0A1W2WPB5 B0WHH5 A0A0K8R750 V5GG89 J3JVF0 G1KD11 A0A131XZ82 A0A023FY01 A0A0C9S225 A0A0P4VUS9 R4FJZ3 A0A1E1X9L8 T1KR88 B7QIG2 H2THT1 U5EKG9 A0A131XDM1 V8P3L0 A0A023GGU3 H9H6Z7 G3MPQ8 A0A131YIZ8 A0A182GCZ4 A0A1W4UVF1 A0A212FF79 T1D9U4 A0A224Z5T5 A0A023FHX0 A0A1D5PQW2 E2A6X8 H2RJ98 A0A023ELB7 A0A195C1U8 A0A151NHF5 A0A1J1IHA7 A0A182GNB4 A0A1V4JB57 A0A0N0PDS2 A0A1W7REV7 A0A0F7Z640 Q0IEM8 A0A183QQP1 A0A218UZV2 A0A293LZ40 Q6GNE0 J3S998 U3FWE0 A0A2P4W7H3 A0A194QAS7 A0A2P6K547 A0A182LXJ3 G1NY02 A0A1B0CL90 A0A3Q7RBJ5 A0A2Y9KEQ3 H0ZHY9 A0A1L8GGM9 A0A2D4M6K7 A0A287D5V8 A0A1S4FMN1
PDB
6CFS
E-value=1.85521e-77,
Score=733
Ontologies
PATHWAY
GO
PANTHER
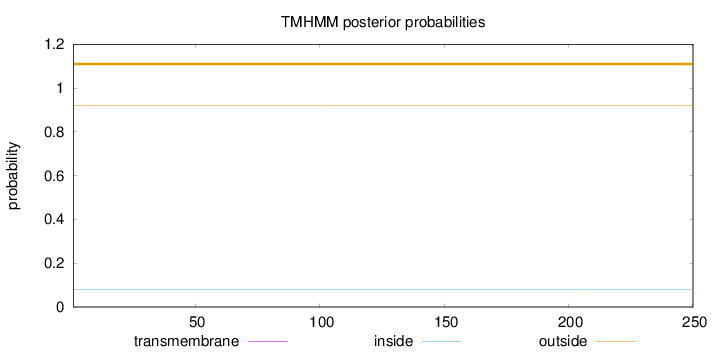
Topology
Subcellular location
Cytoplasm
Length:
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00334
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08088
outside
1 - 250
Population Genetic Test Statistics
Pi
132.27773
Theta
175.614761
Tajima's D
-0.624535
CLR
160.040095
CSRT
0.214239288035598
Interpretation
Uncertain
